Merge Geovars#
Merge all the geovariables and write results toCSV file and STATA Author: Dunstan Matekenya
Affiliation: DECAT, The World Bank Group
Date: August 4, 2023
import warnings
warnings.simplefilter(action='ignore')
import sys
from pathlib import Path
from IPython.display import Image, display
from matplotlib import pyplot as plt
import geopandas as gpd
import numpy as np
import pandas as pd
pd.set_option('display.float_format', lambda x: '%.3f' % x)
spanon = Path.cwd().parents[2].joinpath("Spatial-Anonymization/src/")
sys.path.insert(0, str(spanon))
from spatial_anonymization import utils
from spatial_anonymization import point_displacement
import rasterio
from rasterstats import gen_zonal_stats
# =============================
# BASE WORKING DIRS
# ============================
DIR_DATA = Path.cwd().parents[1].joinpath('data')
DIR_SPATIAL = Path.cwd().parents[1].joinpath('data', 'TZA', 'spatial')
DIR_NPS_W4 = DIR_DATA.joinpath('TZA', 'surveys', 'NPS_w4')
DIR_NPS_W5 = DIR_DATA.joinpath('TZA', 'surveys', 'NPS_w5')
DIR_OUTPUTS = DIR_NPS_W5.joinpath('geovars', 'puf')
# =============================
# SURVEY COORDINATES
# ============================
# WAVE-4 coordinates-public
FILE_PUB_HH_W4 = DIR_NPS_W4.joinpath('geovars', 'npsy4.ea.offset.dta')
FILE_W4_TMP = DIR_NPS_W4.joinpath('geovars', 'hh_sec_a.dta')
# WAVE-4 coordinates-raw
FILE_HH_W4 = DIR_NPS_W4.joinpath('geovars', 'HH_SEC_GPS.dta')
# WAVE-5 coordinates-raw
FILE_HH_W5 = DIR_NPS_W5.joinpath('survey', 'HH_SEC_GPS.dta')
# WAVE-5 FROM MICRODATA-LIB
FILE_HH_W5_MICROLIB = DIR_NPS_W5.joinpath( 'survey', 'from-microdata-lib', 'hh_sec_a.dta')
# ========================
# ADM FILES
# ========================
# Admin regions (adm2) shapefile
FILE_ADM3 = DIR_SPATIAL.joinpath("wards2012", "TZwards-wgs84.shp")
# ============================
# OTHER FILES
# ==================
FILE_POP20 = DIR_DATA.joinpath('WORLDPOP', 'tza', 'tza_ppp_2020.tif')
FILE_WATER_BODIES = DIR_SPATIAL.joinpath('waterbodies_wgs84.shp')
# ====================
# INPUT DATA SPECS
# =====================
# Cols for input coordinates file
LAT_W5 = 'lat_w5'
LON_W5 = 'lon_w5'
LAT_PUB_W4 = 'lat_pub_w4'
LON_PUB_W4 = 'lon_pub_w4'
LAT_PUB_W5 = 'lat_pub_w5'
LON_PUB_W5 = 'lon_pub_w5'
LAT_W4 = 'lat_w4'
LON_W4 = 'lon_w4'
LAT_W4_CENTROID = 'lat_cntr_w4'
LON_W4_CENTROID = 'lon_cntr_w4'
LAT_W5_CENTROID = 'lat_cntr_w5'
LON_W5_CENTROID = 'lon_cntr_w5'
HHID_W5 = 'y5_hhid'
HHID_W4 = 'y4_hhid'
CLUSTID_W4 = 'y4_clusterid'
CLUSTID_W5 = 'y5_clusterid'
URB_RURAL = 'y5_clustertype'
Preprocess Data#
Notes#
Survey identifiers. Prefix Y and W are used interchangeably to refer to survey NPS survey wave
Y4 was conducted in 2014-2015
Y5 was conducted in 2020-2021
HH refers to Household while HHs is plural
Binary variables. For most cases, 1->positive (yes) while 0->negative(no)
Helper variables for coordinates review and mover detection#
The following variables were generated.
Y4 and Y5 Cluster centroids.
'lat_cntr_w4', 'lon_cntr_w4', 'lat_cntr_w5', 'lon_cntr_w5'. Simply means of the coordinates when grouped by clusterid. [Exclude some HHs which are too far (e.g., 10km) from the centroid]Distance to Y4 households [dist_hh_y4]. Calculated for all household coordinates in Y5, this is distance to corresponding Y4 coordinates and so we expect ~ 0 for HHs which haven’t moved.
Distance to nearest clucter centroid [nearest_y4centroid]. Note that the cluster centroids are defined as in 1 above. This is also calculated for all HHs. For booster sample HHs, we assign them cluster based on nearest centroid.
Nearest cluster [nearest_clusterid_y4]. Captures HH nearest based on HH coordinates and clusters centroids. It can be used to compare this clusterid with assigned cluster to check reliability of GPS coordinates.
assigned_vs_nearest_clustid. Compares whether the assigned clusterid is same as the nearest clusterid (as defined in 4 above)
Y4 vs. Y5 clusterid[same_clust].. Check whether HH has same clusterid in Y4 and Y5 again for sanity checks and mover detection.
Imputed coordinates[y4_imputed_coords/y5_imputed_coords]. For data provenance purpose, indicate which HHs are using imputed coordinates.
Additional HHs in Y5 [y5_add_hhs]. For a Y4 HH, this variable indicates how many additional HHs are there in Y5. When
y5_add_hhs = 0it means this household has no split HHs. This maybe useful for mover detection.
Imputing missing coordinates#
Y4 missing coordinates are replaced with Y4 cluster centroids
lat_cntr_w4', lon_cntr_w4Y5 missing coordinates are replaced with Y5 cluster centroids
lat_cntr_w5', lon_cntr_w5
Mover detection notes#
Variables for mover assignment#
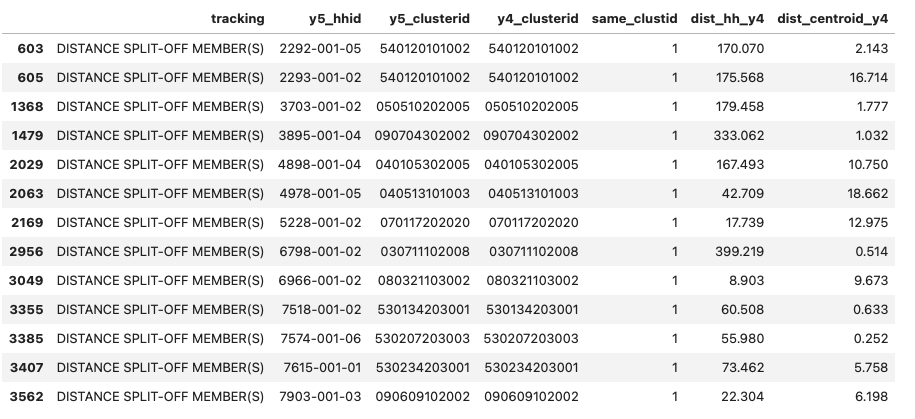
Clusterid for mover HHs. Its noted that mover households were not assigned a new clusterid (see screesnhot below) even though they moved as evidenced by variable
same_clustDistance threshold for mover. If a Y5 HHs is 5km or more from Y4 HHs and/or Y4-clustercentroid, then its a mover.
Variables for detecting movers.
Distance variables:
dist_hh_y4andnearest_y4centroidwhere the distance threshold is applied.Survey tracking status. The survey has variable tracking with the following possible values:
‘ORIGINAL’, ‘LOCAL SPLIT-OFF MEMBER(S)’, ‘DISTANCE SPLIT-OFF MEMBER(S)’, ‘DISTANCE HOUSEHOLD’, ‘BOOSTER SAMPLE (NEW HOUSEHOLD)’
assigned_vs_nearest_clustid - Thiis is used to determine how much to trust/not trust coordinates
imputed_coordinates - Again this variable is used to assign level of trust
Mover status assignment#
Please see function assign_mover_status(row). However, the summary process is shown in screenshot below:
The output variable mover has 4 possible integer values as follows:
3-certain this is amover, 2-mover but with reduced confidence, 1-mover but with reduced confidence, 0- Non-mover For now, categories 1 and above are considered movers. However, category-1 may need review to decide whether to trust the HHs coordinates or not.
display(Image(filename='../../docs/images/tza-npsy5-mover-assignment.png'))

display(Image(filename='../../docs/images/tza-npsy5-movers-clusterid.png'))
def assign_mover_status(row):
# ================================================
# COLLECT VARIABLES TO USE TO DECIDE MOVER STATUS
# ================================================
# Distance from y5 to y4 HH rounded to integer
dist_y4 = round(row['dist_hh_y4'], 0)
# Distance from y5 to y4 centroid
dist_y4centroid = round(row['dist_centroid_y4'], 0)
# Tracking status
tracking_status = row['tracking']
# Whether nearest cluster is same as assigned cluster
y4_clust_agreement = row['assigned_vs_nearest_clustid']
# If tracking_status is any of this, we consider HH a mover
movers = ['LOCAL SPLIT-OFF MEMBER(S)', 'DISTANCE SPLIT-OFF MEMBER(S)', 'DISTANCE HOUSEHOLD']
# Mover status based on "tracking_status" variable
# This is given first precedence
if tracking_status in movers:
survey_mover_assignment = True
else:
survey_mover_assignment = False
# =======================
# ASSIGN MOVER STATUS
# =======================
# Scenario-1: certain this is a mover if distance to y4-hh, distance to y4-centroid and
# mover_survey_assignment1 are true
if (dist_y4 > 5) and (dist_y4centroid > 5) and (survey_mover_assignment):
return 3
# Scenario-2: certain this is a mover if distance to y4-hh, distance to y4-centroid and
# mover_survey_assignment1 are true
if (dist_y4 > 5 and survey_mover_assignment) or (dist_y4centroid > 5 and survey_mover_assignment):
return 3
# Scenario-3: certain mover if both y4-hh and y4-centroid aare above 5
if (dist_y4 > 5) and (dist_y4centroid > 5):
return 3
# Scenario-4: certain mover if distance to y4-hh, distance to y4-centroid and
# mover_survey_assignment1 are true
if dist_y4 > 5:
if (y4_clust_agreement == 1) and (row['y5_imputed_coords'] == 0 or row['y4_imputed_coords'] == 0):
return 2
else:
return 1
return 0
def load_and_preprocess_w4_files():
"""
Do the following preprocessing
1. Merge all multiple files into 1
2. Check for extreme and/or outlier coordinates
2. Replace missing values (if any) with cluster centroids
"""
# ===========================================
# LOAD ALL THE FILES WE NEED
# ===========================================
df_w4_pub = pd.read_stata(FILE_PUB_HH_W4)
df_w4_raw = pd.read_stata(FILE_HH_W4)
df_w4_tmp = pd.read_stata(FILE_W4_TMP)
df_w4_tmp['clusterid'] = df_w4_tmp['clusterid'].apply(lambda x: str(int(x)))
df_w4_pub['clusterid'] = df_w4_pub['clusterid'].apply(lambda x: str(int(x)))
tmp = df_w4_tmp.merge(df_w4_pub[['clusterid', 'lat_modified', 'lon_modified']], on='clusterid')
assert len(tmp) == len(df_w4_raw)
df_w4 = df_w4_raw.merge(tmp[['clusterid', 'clustertype', 'lat_modified', 'lon_modified', 'y4_hhid']], on=HHID_W4)
# =====================
# DO SOME CLEAN-UP
# =====================
# Change DMS to decimal degrees in df_w4_raw
df_w4[LAT_W4] = df_w4.apply(lambda x: -round(x['hh_u1_lat_d'] + x['hh_u1_lat_md']/60, 6), axis=1)
df_w4[LON_W4] = df_w4.apply(lambda x: round(x['hh_u1_lon_d'] + x['hh_u1_lon_md']/60, 6), axis=1)
# Rename columns and keep only those we need
df_w4.rename(columns={'lat_modified':LAT_PUB_W4, 'lon_modified':LON_PUB_W4, 'clusterid':CLUSTID_W4, 'clustertype': 'y4_clustertype'}, inplace=True)
df_w4 = df_w4[['y4_hhid', CLUSTID_W4, 'y4_clustertype', 'lat_pub_w4', 'lon_pub_w4', 'lat_w4', 'lon_w4']]
# Format HHID and clusterid
# Ensure HHID is a string
df_w4[HHID_W4] = df_w4[HHID_W4].astype(str)
# Padd zero to clusterid
df_w4[CLUSTID_W4] = df_w4[CLUSTID_W4].apply(lambda x: '0{}'.format(x) if len(str(x))==11 else str(x))
# ========================================
# DEAL WITH MISSING AND EXTREME VALUES
# ========================================
lon_missing = df_w4[LON_W4].isna().sum()
lat_missing = df_w4[LAT_W4].isna().sum()
# Print missing values
print('='*35)
print('Y4-MISSING COORDINATES REPORT')
print('='*35)
print(df_w4[[LON_W4, LAT_W4]].isna().sum())
print('-'*35)
print(df_w4[df_w4[LON_W4].isna()][['y4_hhid', CLUSTID_W4, 'lat_w4', 'lon_w4']])
# Print Lat and Lon ranges
print()
print('='*35)
print('Y4-CHECKING RANGE OF COORDINATES')
print('='*35)
print(df_w4[[LAT_W4, LON_W4]].describe())
print('-'*35)
# ====================================
# ADD WAVE-4 CLUSTER CENTROIDS
# ====================================
# add wave-4 coordinates cluster centroids
df_clust = df_w4.groupby(CLUSTID_W4).agg({LAT_W4: 'mean', LON_W4:'mean'}).reset_index()
df_clust.rename(columns = {LAT_W4: LAT_W4_CENTROID, LON_W4: LON_W4_CENTROID}, inplace=True)
df_w4 = df_w4.merge(df_clust, on=CLUSTID_W4, how='left')
assert df_w4[LAT_W4_CENTROID].nunique() == len(df_w4_pub)
# Save cluster centroids as shapefile
gdf_clust = gpd.GeoDataFrame(df_clust, geometry=gpd.points_from_xy(df_clust[LON_W4_CENTROID], df_clust[LAT_W4_CENTROID]),
crs="EPSG:4326")
gdf_clust.to_file(DIR_SPATIAL.joinpath('y4-clust-centroids.shp'), driver='ESRI Shapefile')
print('='*62)
print('Y4: REPLACING MISSING COORDS WITH CLUSTER CENTROID IN {} HHs'.format(lon_missing))
print('='*62)
df_w4['y4_imputed_coords'] = df_w4[LAT_W4].apply(lambda x: 1 if np.isnan(x) else 0)
df_w4[LAT_W4] = df_w4.apply(lambda x: x[LAT_W4_CENTROID] if np.isnan(x[LAT_W4]) else x[LAT_W4], axis=1)
df_w4[LON_W4] = df_w4.apply(lambda x: x[LON_W4_CENTROID] if np.isnan(x[LON_W4]) else x[LON_W4], axis=1)
print(df_w4.query('y4_imputed_coords == 1')[['y4_hhid', CLUSTID_W4, 'lat_w4', 'lon_w4',LON_W4_CENTROID, LAT_W4_CENTROID]])
assert df_w4[LON_W4].isna().sum() == 0
assert df_w4[LAT_W4].isna().sum() == 0
assert df_w4.y4_imputed_coords.value_counts()[1] == lon_missing, 'FILLED VLAUES SHOULD MATCH THOSE MISSING'
assert df_w4.y4_imputed_coords.value_counts()[1] == lat_missing, 'FILLED VLAUES SHOULD MATCH THOSE MISSING'
return df_w4
def load_and_preprocess_w5_files():
"""
Do the following preprocessing
1. Merge all multiple files into 1
2. Check for extreme and/or outlier coordinates
2. Replace missing values (if any) with cluster centroids
"""
# HHS with raw coordinates
df_w5 = pd.read_stata(FILE_HH_W5)
# HHS with cluster id
df_w5_pub = pd.read_stata(FILE_HH_W5_MICROLIB)
# =====================
# DO SOME CLEAN-UP
# =====================
# Format clusterid: Padd zero to clusterid from y4 and remove "-" from y5 cluster
df_w5_pub[CLUSTID_W5] = df_w5_pub['y5_cluster'].apply(lambda x: x.replace("-", ""))
assert df_w5_pub[CLUSTID_W5].isna().sum() == 0, 'THERE SHOULD BE NO NULLS FOR CLUSTERID'
# Merge the two dataframes
df_w5 = df_w5_pub[['y5_hhid', 'y5_rural', CLUSTID_W5]].merge(df_w5[['y5_hhid', 'hh_gps__Latitude',
'hh_gps__Longitude', 'hh_a10', 'tracking_class', 'booster']], on=HHID_W5)
df_w5.rename(columns={'hh_gps__Longitude':LON_W5, 'hh_gps__Latitude':LAT_W5, 'y5_rural': URB_RURAL,
'tracking_class': 'tracking', 'booster': 'booster'}, inplace=True)
# Add y4-hhid
df_w5[HHID_W4] = df_w5[HHID_W5].apply(lambda x: x[:-3])
# Ensure clustertype/urban/rural isnt doesnt have nulls
assert df_w5[URB_RURAL].isna().sum() == 0,' URBAN-RURAL COLUMN SHOULDNT HAVE NULLS'
# ========================================
# DEAL WITH MISSING AND EXTREME VALUES
# ========================================
lon_missing = df_w5[LON_W5].isna().sum()
lat_missing = df_w5[LAT_W5].isna().sum()
# Print missing values
print('='*35)
print('Y5-MISSING COORDINATES REPORT')
print('='*35)
print(df_w5[[LON_W5, LAT_W5]].isna().sum())
print('-'*35)
print(df_w5[df_w5[LON_W5].isna()][[HHID_W5, CLUSTID_W5, LAT_W5, LON_W5, 'tracking']])
# Print Lat and Lon ranges
print()
print('='*35)
print('Y5-CHECKING RANGE OF COORDINATES')
print('='*35)
print(df_w5[[LAT_W5, LON_W5]].describe())
print('-'*35)
# ====================================
# ADD WAVE-5 CLUSTER CENTROIDS
# ====================================
# add wave-4 coordinates cluster centroids
df_clust = df_w5.groupby(CLUSTID_W5).agg({LAT_W5: 'mean', LON_W5:'mean'}).reset_index()
df_clust.rename(columns = {LAT_W5: LAT_W5_CENTROID, LON_W5: LON_W5_CENTROID}, inplace=True)
df_w5 = df_w5.merge(df_clust, on=CLUSTID_W5, how='left')
assert df_w5[LAT_W5_CENTROID].nunique() == len(df_clust)
# Save cluster centroids as shapefile
gdf_clust = gpd.GeoDataFrame(df_clust, geometry=gpd.points_from_xy(df_clust[LON_W5_CENTROID], df_clust[LAT_W5_CENTROID]),
crs="EPSG:4326")
gdf_clust.to_file(DIR_SPATIAL.joinpath('y5-clust-centroids.shp'), driver='ESRI Shapefile')
print('='*62)
print('Y5: REPLACING MISSING COORDS WITH CLUSTER CENTROID IN {} HHs'.format(lon_missing))
print('='*62)
df_w5['y5_imputed_coords'] = df_w5[LAT_W5].apply(lambda x: 1 if np.isnan(x) else 0)
df_w5[LAT_W5] = df_w5.apply(lambda x: x[LAT_W5_CENTROID] if np.isnan(x[LAT_W5]) else x[LAT_W5], axis=1)
df_w5[LON_W5] = df_w5.apply(lambda x: x[LON_W5_CENTROID] if np.isnan(x[LON_W5]) else x[LON_W5], axis=1)
print(df_w5.query('y5_imputed_coords == 1')[['y5_hhid', CLUSTID_W5, 'lat_w5', 'lon_w5',LON_W5_CENTROID, LAT_W5_CENTROID]])
assert df_w5[LON_W5].isna().sum() == 0
assert df_w5[LAT_W5].isna().sum() == 0
assert df_w5.y5_imputed_coords.value_counts()[1] == lon_missing, 'FILLED VLAUES SHOULD MATCH THOSE MISSING'
assert df_w5.y5_imputed_coords.value_counts()[1] == lat_missing, 'FILLED VLAUES SHOULD MATCH THOSE MISSING'
return df_w5
def preprocess_coords(df_w4, df_w5):
"""
Perform the following preprocessing steps:
1. Merge w4 and w5
2. Add distance variables between w4 and w5 panel households (movers and non-movers)
3. Add distance to nearest household for booster samples
4. Assign mover status
"""
# =====================
# MERGE W5 WITH W4
# =====================
df_w5w4 = df_w5.merge(df_w4[['y4_hhid', CLUSTID_W4, 'lat_pub_w4', 'lon_pub_w4', 'y4_clustertype',
'lat_w4', 'lon_w4', 'lat_cntr_w4', 'lon_cntr_w4', 'y4_imputed_coords']], on=HHID_W4, how='left', indicator=True)
df_w5_booster_hhs = df_w5.query('booster == "New (booster) sample"')
print(df_w5w4._merge.value_counts())
assert len(df_w5_booster_hhs) == df_w5w4._merge.value_counts()['left_only'], 'MISMATCHED HHS SHOULD BE SAME AS BOOSTER HHS'
df_w5 = df_w5w4.drop(columns=['_merge'])
# =========================
# CONVERT VARS TO NUMERIC
# ========================
df_w5['booster'] = df_w5.booster.map({'Panel household': 0, 'New (booster) sample': 1})
df_w5 = df_w5[[HHID_W4, HHID_W5, CLUSTID_W5, CLUSTID_W4, URB_RURAL, 'y4_clustertype', 'lat_w5', 'lon_w5',
'lat_pub_w4', 'lon_pub_w4', 'lat_w4', 'lon_w4', LON_W5_CENTROID,
LAT_W5_CENTROID, 'tracking', 'booster', 'y4_imputed_coords', 'y5_imputed_coords']]
# =================================
# ADD DISTANCE VARS
# =================================
# Cluster level dataframe
df_clust_y4 = df_w4[[CLUSTID_W4, LON_W4_CENTROID, LAT_W4_CENTROID]].drop_duplicates(subset=[CLUSTID_W4])
assert len(df_clust_y4) == 419, 'THERE SHOULD BE 419 CLUSTERS'
# Distance to nearest y4-cluster centroid
df_w5['dist_centroid_y4'] = df_w5.apply(lambda x: utils.distance_to_points(
df_clust_y4, (x[LAT_W5],x[LON_W5]), lon_col=LON_W4_CENTROID,lat_col=LAT_W4_CENTROID, output='nearest'), axis=1)
# Nearest y4 clusterid based on distance to nearest centroid
df_w5['nearest_clusterid_y4'] = df_w5.apply(lambda x: utils.distance_to_points(
df_clust_y4, (x[LAT_W5],x[LON_W5]), lon_col=LON_W4_CENTROID,lat_col=LAT_W4_CENTROID, id_col=CLUSTID_W4, output='nearest_id'), axis=1)
# Distance from y5 to corresponding y-4 hh
df_w5['dist_hh_y4'] = df_w5.apply(lambda x: utils.distance_between_points((x[LON_W4], x[LAT_W4]), (x[LON_W5], x[LAT_W5])) if x['booster'] == 0 else np.NaN, axis=1)
# =================================
# ADD MOVER CHECKER VARS
# =================================
df_w5['same_clustid'] = df_w5.apply(lambda x: 1 if x[CLUSTID_W5] == x[ CLUSTID_W4] else 0, axis=1)
df_w5['assigned_vs_nearest_clustid'] = df_w5.apply(lambda x: 1 if x[CLUSTID_W4] == x['nearest_clusterid_y4'] else 0, axis=1)
df_w5_panel = df_w5.query('booster == 0')
df_hh_cnt = pd.DataFrame(df_w5_panel.groupby(HHID_W4).size()).rename(columns={0:'y5_add_hhs'})
df_panel = df_w5_panel.merge(df_hh_cnt, on=HHID_W4)
df_panel['y5_add_hhs'] = df_panel['y5_add_hhs'] - 1
assert len(df_panel) == len(df_w5_panel)
df_w5_booster = df_w5.query('booster == 1')
# ==================================
# ADD MOVER STATUS TO PANEL HHS
# ==================================
df_panel['mover'] = df_panel.apply(lambda x: assign_mover_status(x), axis=1)
assert df_panel['mover'].value_counts().sum() == len(df_panel), 'MOVER VARIABLE SHOULD HAVE NO NULLS'
return df_panel, df_w5_booster
print('+'*40)
print('PROCESSING Y4 COORDINATES')
print('+'*40, '\n')
df_y4 = load_and_preprocess_w4_files()
print()
print('+'*40)
print('PROCESSING Y5 COORDINATES')
print('+'*40, '\n')
df_y5 = load_and_preprocess_w5_files()
df_w5_panel, df_w5_booster = preprocess_coords(df_w4=df_y4, df_w5=df_y5)
++++++++++++++++++++++++++++++++++++++++
PROCESSING Y4 COORDINATES
++++++++++++++++++++++++++++++++++++++++
===================================
Y4-MISSING COORDINATES REPORT
===================================
lon_w4 16
lat_w4 16
dtype: int64
-----------------------------------
y4_hhid y4_clusterid lat_w4 lon_w4
181 1425-001 070205202037 NaN NaN
213 1499-001 070219204037 NaN NaN
1600 4800-001 140313103006 NaN NaN
1621 4843-001 140515101008 NaN NaN
1664 4952-001 040403201002 NaN NaN
1971 5696-001 070323207002 NaN NaN
2010 5790-001 010410102002 NaN NaN
2067 5926-001 250413304329 NaN NaN
2107 6019-001 110305210004 NaN NaN
2385 6683-001 030217102005 NaN NaN
2514 6988-001 080510101004 NaN NaN
2600 7194-001 120210101006 NaN NaN
3027 8204-001 060508301305 NaN NaN
3120 8429-001 170428102003 NaN NaN
3155 8508-001 240121104003 NaN NaN
3301 8853-001 140714103001 NaN NaN
===================================
Y4-CHECKING RANGE OF COORDINATES
===================================
lat_w4 lon_w4
count 3336.000 3336.000
mean -5.730 36.158
std 2.453 3.120
min -11.355 29.604
25% -6.890 33.373
50% -6.095 36.850
75% -3.616 39.221
max -1.118 40.453
-----------------------------------
==============================================================
Y4: REPLACING MISSING COORDS WITH CLUSTER CENTROID IN 16 HHs
==============================================================
y4_hhid y4_clusterid lat_w4 lon_w4 lon_cntr_w4 lat_cntr_w4
181 1425-001 070205202037 -6.854 39.147 39.147 -6.854
213 1499-001 070219204037 -6.867 39.226 39.226 -6.867
1600 4800-001 140313103006 -4.944 32.984 32.984 -4.944
1621 4843-001 140515101008 -5.369 32.558 32.558 -5.369
1664 4952-001 040403201002 -5.083 39.073 39.073 -5.083
1971 5696-001 070323207002 -7.108 39.529 39.529 -7.108
2010 5790-001 010410102002 -6.150 36.219 36.219 -6.150
2067 5926-001 250413304329 -3.462 31.887 31.887 -3.462
2107 6019-001 110305210004 -7.794 35.707 35.707 -7.794
2385 6683-001 030217102005 -3.912 37.652 37.652 -3.912
2514 6988-001 080510101004 -10.116 38.927 38.927 -10.116
2600 7194-001 120210101006 -8.950 33.345 33.345 -8.950
3027 8204-001 060508301305 -7.730 38.939 38.939 -7.730
3120 8429-001 170428102003 -3.869 32.196 32.196 -3.869
3155 8508-001 240121104003 -2.458 34.143 34.143 -2.458
3301 8853-001 140714103001 -4.422 32.276 32.276 -4.422
++++++++++++++++++++++++++++++++++++++++
PROCESSING Y5 COORDINATES
++++++++++++++++++++++++++++++++++++++++
===================================
Y5-MISSING COORDINATES REPORT
===================================
lon_w5 7
lat_w5 7
dtype: int64
-----------------------------------
y5_hhid y5_clusterid lat_w5 lon_w5 tracking
988 2977-001-01 210326202001 NaN NaN LOCAL SPLIT-OFF MEMBER(S)
1553 4043-001-01 190517103006 NaN NaN ORIGINAL
1614 4173-001-01 060210305307 NaN NaN ORIGINAL
2134 5143-001-01 070108204015 NaN NaN ORIGINAL
2270 5475-001-01 070219202045 NaN NaN ORIGINAL
2299 5543-001-02 070226204011 NaN NaN LOCAL SPLIT-OFF MEMBER(S)
2602 6150-001-02 180334302009 NaN NaN ORIGINAL
===================================
Y5-CHECKING RANGE OF COORDINATES
===================================
lat_w5 lon_w5
count 4702.000 4702.000
mean -5.684 35.988
std 2.446 3.028
min -11.476 29.604
25% -6.901 33.340
50% -5.955 36.217
75% -3.437 39.194
max -1.057 40.283
-----------------------------------
==============================================================
Y5: REPLACING MISSING COORDS WITH CLUSTER CENTROID IN 7 HHs
==============================================================
y5_hhid y5_clusterid lat_w5 lon_w5 lon_cntr_w5 lat_cntr_w5
988 2977-001-01 210326202001 -4.623 35.656 35.656 -4.623
1553 4043-001-01 190517103006 -2.379 32.330 32.330 -2.379
1614 4173-001-01 060210305307 -6.721 38.742 38.742 -6.721
2134 5143-001-01 070108204015 -6.800 39.233 39.233 -6.800
2270 5475-001-01 070219202045 -6.901 39.210 39.210 -6.901
2299 5543-001-02 070226204011 -6.895 39.244 39.244 -6.895
2602 6150-001-02 180334302009 -1.743 31.563 31.563 -1.743
both 4164
left_only 545
right_only 0
Name: _merge, dtype: int64
df_w5_panel.assigned_vs_nearest_clustid.value_counts()
1 3373
0 791
Name: assigned_vs_nearest_clustid, dtype: int64
df_w5_panel.query('mover == 1')[['y4_hhid', 'y5_hhid', 'y5_clusterid', 'y4_clusterid', 'y5_clustertype',
'y4_clustertype', 'lat_w5', 'lon_w5', 'lat_pub_w4', 'lon_pub_w4',
'lat_w4', 'lon_w4', 'lon_cntr_w5', 'lat_cntr_w5', 'tracking', 'booster',
'y4_imputed_coords', 'y5_imputed_coords', 'dist_centroid_y4',
'nearest_clusterid_y4', 'dist_hh_y4', 'same_clustid',
'assigned_vs_nearest_clustid', 'y5_add_hhs', 'mover']]
Index(['y4_hhid', 'y5_hhid', 'y5_clusterid', 'y4_clusterid', 'y5_clustertype',
'y4_clustertype', 'lat_w5', 'lon_w5', 'lat_pub_w4', 'lon_pub_w4',
'lat_w4', 'lon_w4', 'lon_cntr_w5', 'lat_cntr_w5', 'tracking', 'booster',
'y4_imputed_coords', 'y5_imputed_coords', 'dist_centroid_y4',
'nearest_clusterid_y4', 'dist_hh_y4', 'same_clustid',
'assigned_vs_nearest_clustid', 'y5_add_hhs', 'mover'],
dtype='object')
df_w5_panel.
Anonymize coordinates#
Perform anonymzation as follows:
Non-movers. No anonymization required, assign y4 public coordinates
Booster sample. Apply anonymization as in a regular sample.
Movers. Apply anonymization at household level.
TO-DO
For (2) and (3) above, generate ri
Apply anonymization to the booster sample and movers#
# =====================
# SETUP PARAMS
# ====================
# urb_max_dist
urb_max = 2000
# rur_max_dist
rur_max = 5000
# rur_max_dist_subset
rur_max_subset = 10000
# urb_min_dist
urb_min = 0
# rur_min_dist
rur_min = 0
# apply_min_dist
apply_min_dist = False
# rur_subset_prop = 10%
rur_sub_prop = 0.1
# max_tries
max_tries = 25
# Maximum number of runs for a whole dataset of coordinates
max_runs = 10
# population threshold
pop_thres = 500
# setup the params dict
PARAMS = {'urb_max_dist': urb_max, 'rur_max_dist': rur_max, 'rur_max_dist_subset':rur_max_subset,
'urb_min_dist': urb_min, 'rur_min_dist': rur_min, 'maximum_tries': max_tries,
'apply_min_dist': apply_min_dist, 'rur_sub_prop': rur_sub_prop, 'admin_reg': 'adm3',
'min_pop': pop_thres, 'max_runs': max_runs, 'utm_epsg_code': 32737}
# ===================================
# PREPARE PROCESSING ARGS AND INPUTS
# ==================================
# add EPSG code for geographic CRS if its not
# default to WGS84
GEO_CRS = 4326
URB_RUR = URB_RURAL
RURAL_CODE = 'rural'
# put all input files into a dict
ANON_FILE_INPUTS = {'pop_tif': FILE_POP20, 'shp_waterbodies': FILE_WATER_BODIES,
'shp_adm_bounds': FILE_ADM3}
# =====================================
# APPLY ANONYMIZATION TO BOOSTER SAMPLE
# =====================================
# For this sample, run anonymization at cluster level
# input dataframe
df_input_booster = df_w5_booster[['y5_hhid', 'y5_clusterid', 'y5_clustertype',
LON_W5_CENTROID, LAT_W5_CENTROID]].drop_duplicates(subset=['y5_clusterid'])
ANON_FILE_INPUTS['input_coords'] = df_input_booster.copy(deep=True)
# displacer object
displacer_booster = point_displacement.SimpleDisplacer(params=PARAMS, inputs=ANON_FILE_INPUTS,
output_dir=DIR_OUTPUTS,lon_col=LON_W5_CENTROID,
lat_col=LAT_W5_CENTROID, urb_rur_col=URB_RUR,
unique_id=CLUSTID_W5, rural_code=RURAL_CODE,
geodetic_crs=GEO_CRS)
# Run displacement
out_fpath_booster = DIR_OUTPUTS.joinpath("y5-booster-sample-disp.csv")
print('RUNNING DISPLACEMENT FOR BOOSTER SAMPLE ....')
print('-'*50)
print()
df_disp_booster = displacer_booster.run_displacer(output_file=out_fpath_booster)
RUNNING DISPLACEMENT FOR BOOSTER SAMPLE ....
--------------------------------------------------
==================================================
Running displacement with the following params
==================================================
1.urb_max_dist-2000-Maximum distance(m) for anonymizing region for coordinates in urban areas
2.rur_max_dist-5000-Maximum distance(m) for anonymizing region for coordinates in rural areas
3.rur_max_dist_subset-10000-Maximum distance(m) for anonymizing region for a subset of coordinates in rural areas
4.urb_min_dist-0-Minimum distance(m) from original coordinates in urban areas
5.rur_min_dist-0-Minimum distance(m) from original coordinates in urban areas
6.maximum_tries-25-Number of times to keep trying when geenrated displaced coordinates do not meet requirements
7.apply_min_dist-False-Whether to apply minimum distance or not
8.rur_sub_prop-0.1-Proportion of rural cooridnates to apply large anonymizing zone
9.admin_reg-adm3-Admin region for constraining displaced coordinates
10.min_pop-500-Minimum number of people withing anonymizing region to avoid re-identification
11.max_runs-10-Maximum number of re-runs when some coordinates fail to get generated
12.utm_epsg_code-32737-While some counties have multiple UTM zones, use this as default
13.pop_check-True-Whether generated coordinates meet the minimum population threshold
14.check_waterbodies-True-Check to ensure generated coordinates dont fall on large water bodies
Params file saved to file:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/disp-params-y5-booster-sample-disp.csv
----------------------------------------------------------------------------------------------------
Preparing data for displacement...
Found the following projected CRS: ['32736', '32737']
Adding displacements to 68 points ....
Run-10 ......
1 coords failed to be generated, re-running
Run-9 ......
Saving outputs...
Displacements generated for all coordinates in input
Output CSV file with anonymized coordinates saved to file:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-booster-sample-disp.csv
Processing 68 coordinates with 25 maximum tries per coordinate took 0 minutes
----------------------------------------------------------------------------------------------------
============================================================
Generating summary statistics to check the displacement
============================================================
Urban/rural distribution
-------------------------
2 68
1 0
Name: y5_clustertype, dtype: int64
Stats for distance between original & displaced coords-Urban
--------------------------------------------------------------
stat area value
0 mean urban 0.692
2 median urban 0.517
4 minimum urban 0.006
6 maximum urban 1.891
Stats for distance between original & displaced coords-Rural
--------------------------------------------------------------
stat area value
1 mean rural NaN
3 median rural NaN
5 minimum rural NaN
7 maximum rural NaN
Summary displacement statistics saved to:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-booster-sample-disp-disp-stats.csv
----------------------------------------------------------------------------------------------------
# =====================================
# APPLY ANONYMIZATION TO MOVERS
# =====================================
# For movers, apply anonymization at HH level using HH coordinates
# input dataframe
df_w5_panel_movers = df_w5_panel.query('mover > 0')[
['y5_hhid', 'y4_hhid', 'y5_clusterid', 'y5_clustertype', LAT_W5, LON_W5]]
ANON_FILE_INPUTS['input_coords'] = df_w5_panel_movers.copy(deep=True)
# displacer object
displacer_movers = point_displacement.SimpleDisplacer(params=PARAMS, inputs=ANON_FILE_INPUTS,
output_dir=DIR_OUTPUTS,lon_col=LON_W5,
lat_col=LAT_W5, urb_rur_col=URB_RUR,
unique_id='y5_hhid', rural_code=RURAL_CODE,
geodetic_crs=GEO_CRS)
# Run displacement
out_fpath_movers = DIR_OUTPUTS.joinpath("y5-movers-disp.csv")
print()
print('RUNNING DISPLACEMENT FOR MOVERS ....')
print('-'*60)
print()
df_disp_movers = displacer_movers.run_displacer(output_file=out_fpath_movers)
RUNNING DISPLACEMENT FOR MOVERS ....
------------------------------------------------------------
==================================================
Running displacement with the following params
==================================================
1.urb_max_dist-2000-Maximum distance(m) for anonymizing region for coordinates in urban areas
2.rur_max_dist-5000-Maximum distance(m) for anonymizing region for coordinates in rural areas
3.rur_max_dist_subset-10000-Maximum distance(m) for anonymizing region for a subset of coordinates in rural areas
4.urb_min_dist-0-Minimum distance(m) from original coordinates in urban areas
5.rur_min_dist-0-Minimum distance(m) from original coordinates in urban areas
6.maximum_tries-25-Number of times to keep trying when geenrated displaced coordinates do not meet requirements
7.apply_min_dist-False-Whether to apply minimum distance or not
8.rur_sub_prop-0.1-Proportion of rural cooridnates to apply large anonymizing zone
9.admin_reg-adm3-Admin region for constraining displaced coordinates
10.min_pop-500-Minimum number of people withing anonymizing region to avoid re-identification
11.max_runs-10-Maximum number of re-runs when some coordinates fail to get generated
12.utm_epsg_code-32737-While some counties have multiple UTM zones, use this as default
13.pop_check-True-Whether generated coordinates meet the minimum population threshold
14.check_waterbodies-True-Check to ensure generated coordinates dont fall on large water bodies
Params file saved to file:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/disp-params-y5-movers-disp.csv
----------------------------------------------------------------------------------------------------
Preparing data for displacement...
Found the following projected CRS: ['32737', '32736', '32735']
Adding displacements to 808 points ....
Run-10 ......
9 coords failed to be generated, re-running
Run-9 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 30})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 30})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 30, 'adm': 10})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 30, 'adm': 27})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 30})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 30, 'adm': 25})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 30, 'adm': 16})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 30, 'adm': 6})
8 coords failed to be generated, re-running
Run-8 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 35})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 35})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 35, 'adm': 16})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 35, 'adm': 19})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 35})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 35, 'adm': 17})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 35, 'adm': 16})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 35, 'adm': 13})
8 coords failed to be generated, re-running
Run-7 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 40})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 40})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 40, 'adm': 18})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 40, 'adm': 31})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 40})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 40, 'adm': 37})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 40, 'adm': 15})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 40, 'adm': 10})
8 coords failed to be generated, re-running
Run-6 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 45})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 45})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 45, 'adm': 11})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 45, 'adm': 37})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 45})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 45, 'adm': 24})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 45, 'adm': 26})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 45, 'adm': 15})
8 coords failed to be generated, re-running
Run-5 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 50})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 50})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 50, 'adm': 20})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 50, 'adm': 28})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 50})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 50, 'adm': 32})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 50, 'adm': 18})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 50, 'adm': 10})
8 coords failed to be generated, re-running
Run-4 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 55})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 55})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 55, 'adm': 22})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 55, 'adm': 32})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 55})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 55, 'adm': 30})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 55, 'adm': 20})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 55, 'adm': 11})
8 coords failed to be generated, re-running
Run-3 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 60})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 60})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 60, 'adm': 23})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 60, 'adm': 43})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 60})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 60, 'adm': 46})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 60, 'adm': 25})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 60, 'adm': 15})
8 coords failed to be generated, re-running
Run-2 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 65})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 65})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 65, 'adm': 24})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 65, 'adm': 39})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 65})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 65, 'adm': 44})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 65, 'adm': 27})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 65, 'adm': 15})
8 coords failed to be generated, re-running
Run-1 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 70})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 70})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 70, 'adm': 20})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 70, 'adm': 38})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 70})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 70, 'adm': 44})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 70, 'adm': 25})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 70, 'adm': 8})
8 coords failed to be generated, re-running
Run-0 ......
Failed to generate displacements for : 1725-001-02-38.21352834,-6.77698754
Counter({'pop': 75})
Failed to generate displacements for : 2867-001-01-39.1242328,-8.7715054
Counter({'pop': 75})
Failed to generate displacements for : 4764-001-01-33.43886467,-7.89284325
Counter({'pop': 75, 'adm': 20})
Failed to generate displacements for : 4764-001-03-33.43181982,-7.87788732
Counter({'pop': 75, 'adm': 47})
Failed to generate displacements for : 7293-001-07-34.49006942,-8.51862129
Counter({'pop': 75})
Failed to generate displacements for : 4706-001-01-33.57930825,-6.76978313
Counter({'pop': 75, 'adm': 60})
Failed to generate displacements for : 1988-001-07-34.81879812,-9.55942101
Counter({'pop': 75, 'adm': 24})
Failed to generate displacements for : 1988-001-06-34.81669505,-9.55582022
Counter({'pop': 75, 'adm': 14})
Exhausted number of tries, existing without finishing all coordinates
8 points couldnt be displaced
y5_hhid y5_clustertype lon_w5 lat_w5
720 1725-001-02 2 38.214 -6.777
1043 2867-001-01 2 39.124 -8.772
1549 4764-001-01 1 33.439 -7.893
1549 4764-001-03 1 33.432 -7.878
1706 7293-001-07 2 34.490 -8.519
1814 4706-001-01 1 33.579 -6.770
2943 1988-001-07 2 34.819 -9.559
2943 1988-001-06 2 34.817 -9.556
Saving outputs...
Coordinates with no displacement saved to file:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-movers-disp-failed.csv
Output CSV file with anonymized coordinates saved to file:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-movers-disp.csv
Processing 800 coordinates with 25 maximum tries per coordinate took 18 minutes
----------------------------------------------------------------------------------------------------
============================================================
Generating summary statistics to check the displacement
============================================================
Urban/rural distribution
-------------------------
1 465
2 335
Name: y5_clustertype, dtype: int64
Stats for distance between original & displaced coords-Urban
--------------------------------------------------------------
stat area value
0 mean urban 0.749
2 median urban 0.609
4 minimum urban 0.001
6 maximum urban 1.972
Stats for distance between original & displaced coords-Rural
--------------------------------------------------------------
stat area value
1 mean rural 2.054
3 median rural 1.765
5 minimum rural 0.025
7 maximum rural 8.945
Summary displacement statistics saved to:
DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-movers-disp-disp-stats.csv
----------------------------------------------------------------------------------------------------
# df = pd.read_csv(DIR_OUTPUTS.joinpath('y5-movers-disp-failed.csv'))
Merge back files and create final y5 public coordinates file#
Add final tracking status column to show which households are panel-movers, panel-non-movers and booster
Merge all into a single dataframe
Ensure all households have public coordinates except for those which didnt get anonymized
# =========================================
# RENAME DISP_COORDS TO MATCH WITH COLNAMES
# IN DF_W5_PANEL
# ========================================
df_disp_booster2 = df_disp_booster.rename(columns={'disp_lon': LON_PUB_W5,
'disp_lat': LAT_PUB_W5})[['y5_clusterid',LON_PUB_W5, LAT_PUB_W5]]
df_disp_movers2 = df_disp_movers[['y5_hhid', 'disp_lon','disp_lat']]
# =============================================
# MERGE CLUSTER LEVEL BOOSTER DATAFRAME WITH
# DISPACED COORDINATES WITH HH LEVEL DATAFRAME
# ============================================
df_w5_booster['tracking_final'] = 'booster'
df_w5_booster2 = df_w5_booster.merge(df_disp_booster2, on='y5_clusterid', how='left', indicator=True)
assert df_w5_booster2._merge.value_counts()['both'] == len(df_w5_booster)
df_w5_booster2.drop(columns=['_merge'], inplace=True)
assert df_w5_booster2[LON_PUB_W5].isna().sum() == 0
assert df_w5_booster2[LAT_PUB_W5].isna().sum() == 0
# ========================================
# MERGE MOVERS BACK TO DF_PANEL DATAFRAMES
# ========================================
# Add a variable (tracking_final) to indicate whether each HH
# is mover, non-mover or booster
df_w5_panel['tracking_final'] = df_w5_panel.mover.apply(lambda x: 'mover' if x > 0 else 'non-mover')
# For non-movers, we carry over the Y4 public coordinates
# as Y5 public coordinates
df_w5_panel[LON_PUB_W5] = df_w5_panel.apply(lambda x: x[LON_PUB_W4] if x['tracking_final'] == 'non-mover' else np.NaN, axis=1)
df_w5_panel[LAT_PUB_W5] = df_w5_panel.apply(lambda x: x[LAT_PUB_W4] if x['tracking_final'] == 'non-mover' else np.NaN, axis=1)
# Merge the movers dataframe with displaced coordinates
# with the rest of the panel dataframe
df_w5_panel2 = df_w5_panel.merge(df_disp_movers2, on='y5_hhid', how='left', indicator=True)
assert df_w5_panel2._merge.value_counts()['both'] == len(df_disp_movers2)
df_w5_panel2[LON_PUB_W5] = df_w5_panel2.apply(lambda x: x['disp_lon'] if x['tracking_final'] == 'mover' else x[LON_PUB_W5], axis=1)
df_w5_panel2[LAT_PUB_W5] = df_w5_panel2.apply(lambda x: x['disp_lat'] if x['tracking_final'] == 'mover' else x[LAT_PUB_W5], axis=1)
df_w5_panel2.drop(columns=['_merge', 'disp_lon', 'disp_lat'], inplace=True)
assert df_w5_panel2[LON_PUB_W5].isna().sum() == 8
assert df_w5_panel2[LAT_PUB_W5].isna().sum() == 8
# ========================================
# MERGE ALL DATAFRAMES TOGETHER
# ========================================
# Append booster sample to panel sample
dfw5 = pd.concat([df_w5_panel2, df_w5_booster2], axis=0)
keep_cols = ['y5_hhid', 'y5_clusterid', 'y5_clustertype',
'tracking_final','lon_cntr_w5', 'lat_cntr_w5', LON_W5, LAT_W5,
'lon_pub_w5', 'lat_pub_w5']
dfw5 = dfw5[keep_cols]
dfw5['make_public'] = dfw5.apply(lambda x: 0 if np.isnan(x[LON_PUB_W5]) else 1, axis=1)
assert len(dfw5) == df_w5_panel.shape[0] + df_w5_booster.shape[0]
# Save to CSV
out_csv = DIR_OUTPUTS.joinpath('y5-hhs-pub-coords.csv')
dfw5.to_csv(out_csv, index=False)
LON_W5
PosixPath('/Users/dunstanmatekenya/Library/CloudStorage/OneDrive-WBG/DECAT_HH_Geovariables/data/TZA/surveys/NPS_w5/geovars/puf/y5-hhs-pub-coords.csv')
LON_PUB_W5
'lon_pub_w5'