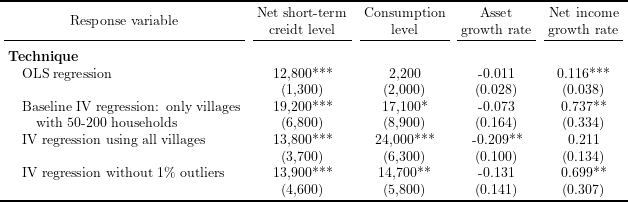
Table 3 in Kaboski and Townsend (2012), “The Impact of Credit on Village Economies”
To read the created .tex file, the following \(\LaTeX\) packages are required:
- float
- pdflscape
- booktabs
- multicol
- makecell
# Install and load packages ---------------
packages <- c(
"tidyverse",
"haven",
"lfe",
"stargazer",
"broom",
"kableExtra",
"scales",
"magrittr"
)
pacman::p_load(packages, character.only = TRUE, install = TRUE)
# Load an example dataset ---------------
data_short <- read_dta("Kaboski2012_replication/AEJApp2009-0115_data/documentation/AnnualData_ShortSample.dta")Data cleaning
table3_outcome_list <- c(
"newst", "tc", "dlgassd", "dnetinc"
)
data_table3 <- data_short %>%
group_by(case_id) %>%
mutate(
farm = ifelse(
!is.na(och),
ifelse((och > -5) & (och <= 15), 1, 0),
0
),
age2h = ageh^2,
vfst = vfst / 10000,
villageyear = floor(case_id / 1000),
invHH = ifelse(year == 1, lead(1 / vHH), 1 / vHH),
invHHim = ifelse(year == 6, invHH, NA),
invHHi = mean(invHHim, na.rm = TRUE),
vHHi = 1 / invHHi,
invHHpvf = invHHi * (year > 5),
invHHtvf1 = invHHi * (year == 6),
invHHtvf2 = invHHi * (year == 7),
dlgassd = log(lead(gassd) / gassd),
dnetinc = ifelse(
is.infinite(log(lead(netinc) / netinc)),
NA,
log(lead(netinc) / netinc)
)
) %>%
ungroup() %>%
filter(year < 8)Create table
# list of functions to filter data to create subsample
table3_filter_list <- list(
function (x) {filter(
x, newst <= 315000,
vHHi < 250, vHHi >= 50, year < 8
)},
function (x) {filter(
x, tc >= 4430, tc <= 433000,
vHHi < 250, vHHi >= 50, year < 8
)},
function (x) {filter(
x, dlgassd >= -2.565758, dlgassd <= 2.494314,
vHHi < 250, vHHi >= 50, year < 8
)},
function (x) {filter(
x, dnetinc >= -2.605709, dnetinc <= 2.848096,
vHHi < 250, vHHi >= 50, year < 8
)}
)
# function for regressions
reg_func <- function(varlist, filterlist, iv, data) {
varlist %>%
enframe("model_no", "outcome") %>%
mutate(filter = filterlist) %>%
mutate(
model = map2(outcome, filter, function(x, y) felm(
formula(
paste0(
x,
"~ as.factor(year) + madult + fadult + kids + maleh + farm + ageh + age2h + educh | ",
"case_id | ", iv, " | villageyear"
)
),
data = y(data)
))
)
}
# functions to arrange regression results for output table
table_arrange <- function(reg_output) {
main_rhs = "`vfst(fit)`"
reg_output %>%
mutate(
coef = map_chr(model, function(x) ifelse(
abs(x$coefficients[main_rhs, 1]) > 50,
comma_format(accuracy = 1e+2)(x$coefficients[main_rhs, 1]),
ifelse(
abs(x$coefficients[main_rhs, 1]) > 5,
comma_format(accuracy = 1e+1)(x$coefficients[main_rhs, 1]),
comma_format(accuracy = 1e-3)(x$coefficients[main_rhs, 1])
)
)),
pval = map_dbl(model, ~ .x$cpval[main_rhs]),
coef = ifelse(
pval < 0.01,
paste0(coef, "***"),
ifelse(
pval < 0.05,
paste0(coef, "**"),
ifelse(
pval < 0.10,
paste0(coef, "*"),
coef
)
)
),
se = paste0(
"(",
map_chr(model, function(x) ifelse(
abs(x$cse[main_rhs]) > 50,
comma_format(accuracy = 1e+2)(x$cse[main_rhs]),
ifelse(
abs(x$cse[main_rhs]) > 5,
comma_format(accuracy = 1e+1)(x$cse[main_rhs]),
comma_format(accuracy = 1e-3)(x$cse[main_rhs])
)
)),
")"
)
) %>%
select(coef, se) %>%
as.matrix() %>%
t()
}
# OLS regression
table3_ols_output <- reg_func(
table3_outcome_list,
replicate(
length(table3_outcome_list),
function(x) {filter(x, vHHi < 250, vHHi >= 50, year < 8)}
),
"(vfst ~ vfst)",
data_table3
) %>%
table_arrange()
# IV regression
table3_baseline_iv_output <- reg_func(
table3_outcome_list,
replicate(
length(table3_outcome_list),
function(x) {filter(x, vHHi < 250, vHHi >= 50, year < 8)}
),
"(vfst ~ invHHtvf1 + invHHtvf2)",
data_table3
) %>%
table_arrange()
# IV regression, all villages
table3_all_iv_output <- reg_func(
table3_outcome_list,
replicate(
length(table3_outcome_list),
function(x) {filter(x, year < 8)}
),
"(vfst ~ invHHtvf1 + invHHtvf2)",
data_table3
) %>%
table_arrange()
# IV regression with truncation
table3_no_outlier_iv_output <- reg_func(
table3_outcome_list,
table3_filter_list,
"(vfst ~ invHHtvf1 + invHHtvf2)",
data_table3
) %>%
table_arrange()
# combine results for output table
c(
"OLS regression", "",
"Baseline IV regression: only villages", "\\hspace{1em}with 50-200 households",
"IV regression using all villages", "",
"IV regression without 1\\% outliers", ""
) %>%
cbind(
rbind(
table3_ols_output,
table3_baseline_iv_output,
table3_all_iv_output,
table3_no_outlier_iv_output
)
) %>%
as_tibble() %>%
mutate_all(linebreak, align = "l") %>%
set_names(NULL) %>%
kable(
"latex", booktabs = TRUE, escape = FALSE,
align = c("l", rep("c", length(table3_outcome_list)))
) %>%
kable_styling(latex_options = "scale_down") %>%
add_header_above(
c(
"Response variable", "Net short-term\ncreidt level",
"Consumption\nlevel", "Asset\ngrowth rate",
"Net income\ngrowth rate"
)
) %>%
pack_rows(index = c(
"Technique" = 8
)) %>%
save_kable("tex/Kaboski2012_table3_replicate.tex")