4. Mali#
Author: Andres Chamorro
Date: March 15, 2023
The objective of this analysis is to construct health facility access indicators over the general population, and across the socioeconomic distribution for a set of GFF countries with available geolocalized facility level information.
Indicators of interest
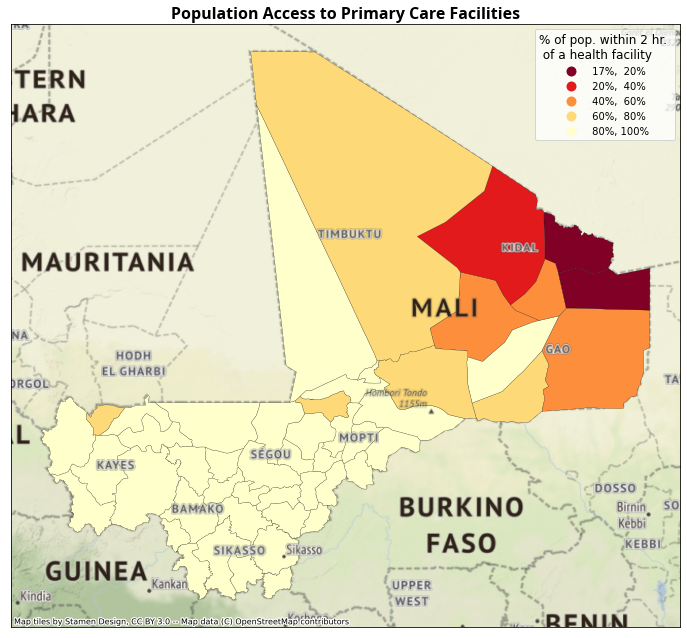
Percentage of population within 2h of driving to the nearest primary care facility (population level, and by SES quintile).
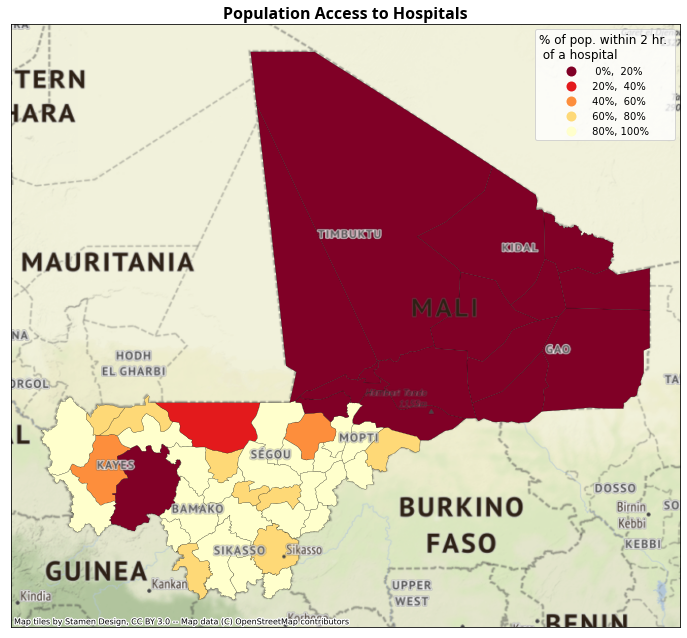
Percentage of population within 2h of driving to the nearest district hospital (population, and by SES quintile).
Percentage of health facilities with direct access to an all season road.
Percentage of health facilities within 2km of an all season road.
Data Sources
Health facilities: National Registry Master List, we geo-coded entries with missing coordinates
Administrative Boundaries: World Bank GAUL Level 1/2
Population 2020: WorldPop 1km Grid Unconstrained
4.1. Data preparation#
This first section conducts some common data preparation tasks:
Import libraries
Load input data (this notebook was ran from the DEC JNB server)
Merge population and friction surface raster data
Prepare origins (population grid) and destinations (health facilities)
Show code cell content
## Define Imports
import os, sys
from os.path import join
from pathlib import Path
import geopandas as gpd
import pandas as pd
import rasterio as rio
import numpy as np
from shapely.geometry import Point
import skimage.graph as graph
from rasterstats import zonal_stats
# for plotting maps
import matplotlib.pyplot as plt
import matplotlib.colors as colors
from matplotlib import colorbar
from rasterio.plot import plotting_extent
import contextily as ctx
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
# for roads
import json
from utm_zone import epsg as epsg_get
# for facebook data
from pyquadkey2 import quadkey
# for graphs
from plotnine import ggplot, aes, geom_col, labs, theme_minimal, theme, element_blank, facet_wrap, scale_y_continuous
from mizani.formatters import percent_format
import seaborn as sns
import matplotlib.ticker as mtick
## TO DO: Work to consolidate functions onto a sinle package/module
sys.path.append('/home/wb514197/Repos/gostrocks/src') # gostrocks is used for some basic raster operations (clip and standardize)
sys.path.append('/home/wb514197/Repos/GOSTNets_Raster/src') # gostnets_raster has functions to work with friction surface
sys.path.append('/home/wb514197/Repos/INFRA_SAP') # only used to save some raster results
sys.path.append('/home/wb514197/Repos/health-equity-diagnostics/src/modules') #
import GOSTRocks.rasterMisc as rMisc
import GOSTNetsRaster.market_access as ma
from infrasap import aggregator
from infrasap import osm_extractor as osm
from utils import download_osm_shapefiles
Define inputs
## health and admin data
iso3 = "MLI"
country = "mali"
geocoding_output = f"{iso3}_geocoding_2.7.23.json"
input_dir = "/home/public/Data/PROJECTS/Health"
out_folder = os.path.join(input_dir, "output", iso3)
if not os.path.exists(out_folder):
os.mkdir(out_folder)
Show code cell content
global_admin = '/home/public/Data/GLOBAL/ADMIN/g2015_0_simplified.shp'
adm0 = gpd.read_file(global_admin)
aoi = adm0.loc[adm0.ISO3166_1_==iso3]
global_admin2 = '/home/public/Data/GLOBAL/ADMIN/Admin2_Polys.shp'
adm2 = gpd.read_file(global_admin2)
adm2 = adm2.loc[adm2.ISO3==iso3].copy()
adm2 = adm2.to_crs("EPSG:4326")
adm2.reset_index(inplace=True)
## Friction Surface and Population
global_friction_surface = "/home/public/Data/GLOBAL/INFRA/FRICTION_2020/2020_motorized_friction_surface.geotiff"
global_population = "/home/public/Data/GLOBAL/Population/WorldPop_PPP_2020/ppp_2020_1km_Aggregated.tif"
inG = rio.open(global_friction_surface)
# Clip the travel raster to AOI
out_travel_surface = os.path.join(out_folder, "travel_surface.tif")
rMisc.clipRaster(inG, aoi, out_travel_surface, crop=True, buff=0.1)
inP = rio.open(global_population)
# Clip the pop raster to AOI
out_pop = os.path.join(out_folder, "WP_2020_1km.tif")
rMisc.clipRaster(inP, aoi, out_pop, crop=True, buff=0.1)
travel_surf = rio.open(out_travel_surface)
pop_surf = rio.open(out_pop)
# standardize so that they have the same number of pixels and dimensions
out_pop_surface_std = os.path.join(out_folder, "WP_2020_1km_STD.tif")
rMisc.standardizeInputRasters(pop_surf, travel_surf, out_pop_surface_std, resampling_type="nearest")
# create a data frame of all points
pop_surf = rio.open(out_pop_surface_std)
pop = pop_surf.read(1, masked=True)
indices = list(np.ndindex(pop.shape))
xys = [Point(pop_surf.xy(ind[0], ind[1])) for ind in indices]
res_df = pd.DataFrame({
'spatial_index': indices,
'xy': xys,
'pop': pop.flatten()
})
res_df['pointid'] = res_df.index
# create MCP object
inG_data = travel_surf.read(1) * 1000 # minutes to travel 1 meter, convert to km
# Correct no data values
inG_data[inG_data < 0] = 9999999999 # untraversable
# inG_data[inG_data < 0] = np.nan
mcp = graph.MCP_Geometric(inG_data)
/home/wb514197/Repos/gostrocks/src/GOSTRocks/rasterMisc.py:73: UserWarning: Geometry is in a geographic CRS. Results from 'buffer' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
/home/wb514197/Repos/gostrocks/src/GOSTRocks/rasterMisc.py:73: UserWarning: Geometry is in a geographic CRS. Results from 'buffer' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
Summary of health facilities by type
Hospital tag was assigned if facility name contains any of the following words
searchfor = ['hop', 'hosp', 'hôp', 'cabinet medical', 'cabinet médical']
Show code cell source
## Destinations
# master = pd.read_csv(join(input_dir, "from_tashrik", "master lists", "Liberia MFL Adjusted.csv"), index_col=0) Médical
master = None
geocoded = gpd.read_file(join(out_folder, geocoding_output))
geocoded = geocoded.loc[~geocoded.geometry.isna()]
geocoded.loc[:, "facilitytype"] = "None"
geocoded.loc[:, "orgunitlevel5_edit"] = geocoded.orgunitlevel5.str.lower()
geocoded.loc[geocoded.orgunitlevel5_edit.str.contains('|'.join(searchfor)), 'facilitytype'] = 'hospital'
health = pd.concat([master, geocoded])
health = health.loc[health.intersects(aoi.unary_union)]
health.reset_index(inplace=True, drop=True)
hospitals = health.loc[health.facilitytype=="hospital"].copy()
geocoded.facilitytype.value_counts()
None 1784
hospital 103
Name: facilitytype, dtype: int64
4.2. Calculate travel time to nearest health facility#
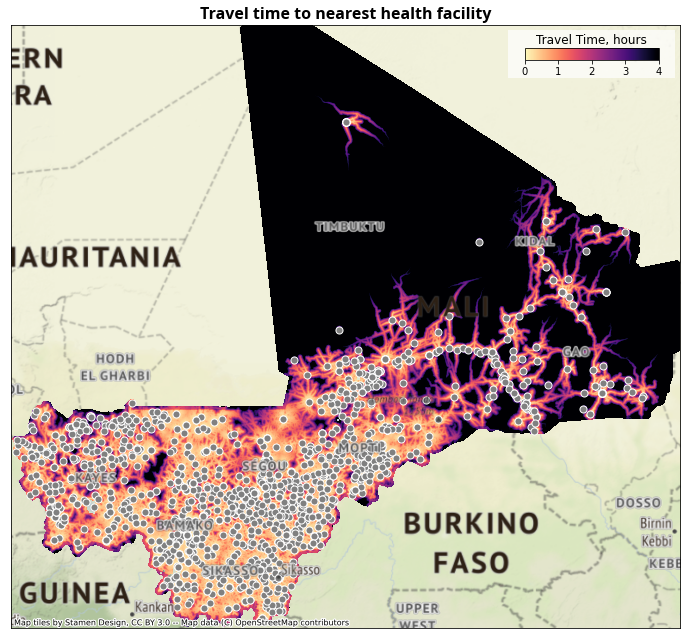
We calculate driving time from every populated place (using a 1 km population grid) to the nearest health facility, using the global friction surface.
Show code cell source
## Calculate travel time to nearest health facility / hospital
res_health = ma.calculate_travel_time(travel_surf, mcp, health)[0]
res_hospital = ma.calculate_travel_time(travel_surf, mcp, hospitals)[0]
res_df.loc[:, f"tt_health"] = res_health.flatten()
res_df.loc[:, f"tt_hospital"] = res_hospital.flatten()
res_df = res_df.loc[res_df['pop']>0].copy()
res_df = res_df.loc[~(res_df['pop'].isna())].copy()
res_df.loc[:,'xy'] = res_df['xy'].apply(Point)
res_gdf = gpd.GeoDataFrame(res_df, geometry='xy', crs='EPSG:4326')
res_gdf.loc[:, 'geometry'] = res_gdf.loc[:, 'xy']
raster_path = out_pop_surface_std
aggregator.rasterize_gdf(res_gdf, 'tt_health', raster_path, os.path.join(out_folder, "tt_health.tif"), nodata=-1)
aggregator.rasterize_gdf(res_gdf, 'tt_hospital', raster_path, os.path.join(out_folder, "tt_hospital.tif"), nodata=-1)
tt_health_rio = rio.open(join(out_folder, "tt_health.tif"))
tt_health = tt_health_rio.read(1, masked=True)
tt_hospital_rio = rio.open(join(out_folder, "tt_hospital.tif"))
tt_hospital = tt_hospital_rio.read(1, masked=True)
tt_health = tt_health/60
tt_hospital = tt_hospital/60
4.2.1. Travel Time Maps#
Show code cell source
figsize = (12,12)
fig, ax = plt.subplots(1, 1, figsize = figsize)
fonttitle = {'fontname':'Open Sans','weight':'bold','size':16}
ax.set_title("Travel time to nearest health facility", fontdict=fonttitle)
ax.get_xaxis().set_visible(False) # plt.axis('off')
ax.get_yaxis().set_visible(False)
vmin = 0
vmax = 4
cmap = 'magma_r'
ext = plotting_extent(tt_health_rio)
im = ax.imshow(tt_health, cmap=cmap, extent=ext, vmin=vmin, vmax=vmax) # norm=colors.PowerNorm(gamma=0.3)
orientation='horizontal'
alpha=1
cbbox = inset_axes(
ax, '25%', '8%', loc = 'upper right'
)
cbbox.set_title('Travel Time, hours', y=1.0, pad=-14)
[cbbox.spines[k].set_visible(False) for k in cbbox.spines]
cbbox.tick_params(
axis='both', which='both',
left=False, top=False, right=False, bottom=False,
labelleft=False, labeltop=False, labelright=False, labelbottom=False
)
cbbox.set_facecolor([1,1,1,0.7])
# cbbox.set_axis_off()
cax = inset_axes(cbbox, '80%', '25%', loc = 'center')
cmap = plt.get_cmap(cmap)
norm = colors.Normalize(vmin=vmin, vmax=vmax)
cb = colorbar.ColorbarBase(
ax=cax, norm=norm, alpha=alpha, cmap=cmap, orientation=orientation,
)
health.plot(ax=ax, facecolor='gray', edgecolor='white', markersize=50, alpha=1)
# ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10)
ctx.add_basemap(ax, source=ctx.providers.Stamen.TerrainLabels, crs='EPSG:4326', zorder=1)
ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10, alpha=0.75)
# plt.tight_layout()
plt.savefig(join(out_folder, "Friction_TT_health.png"), dpi=150, bbox_inches='tight', facecolor='white')
Show code cell source
figsize = (12,12)
fig, ax = plt.subplots(1, 1, figsize = figsize)
fonttitle = {'fontname':'Open Sans','weight':'bold','size':16}
ax.set_title("Travel time to nearest hospital", fontdict=fonttitle)
ax.get_xaxis().set_visible(False) # plt.axis('off')
ax.get_yaxis().set_visible(False)
vmin = 0
vmax = 4
cmap = 'magma_r'
ext = plotting_extent(tt_health_rio)
im = ax.imshow(tt_hospital, cmap=cmap, extent=ext, vmin=vmin, vmax=vmax) # norm=colors.PowerNorm(gamma=0.3)
orientation='horizontal'
alpha=1
cbbox = inset_axes(
ax, '25%', '8%', loc = 'upper right'
)
cbbox.set_title('Travel Time, hours', y=1.0, pad=-14)
[cbbox.spines[k].set_visible(False) for k in cbbox.spines]
cbbox.tick_params(
axis='both', which='both',
left=False, top=False, right=False, bottom=False,
labelleft=False, labeltop=False, labelright=False, labelbottom=False
)
cbbox.set_facecolor([1,1,1,0.7])
cax = inset_axes(cbbox, '80%', '25%', loc = 'center')
cmap = plt.get_cmap(cmap)
norm = colors.Normalize(vmin=vmin, vmax=vmax)
cb = colorbar.ColorbarBase(
ax=cax, norm=norm, alpha=alpha, cmap=cmap, orientation=orientation,
)
hospitals.plot(ax=ax, facecolor='navy', edgecolor='white', markersize=80, alpha=1)
# ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10)
ctx.add_basemap(ax, source=ctx.providers.Stamen.TerrainLabels, crs='EPSG:4326', zorder=1)
ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10, alpha=0.75)
plt.savefig(join(out_folder, "Friction_TT_hospital.png"), dpi=150, bbox_inches='tight', facecolor='white')
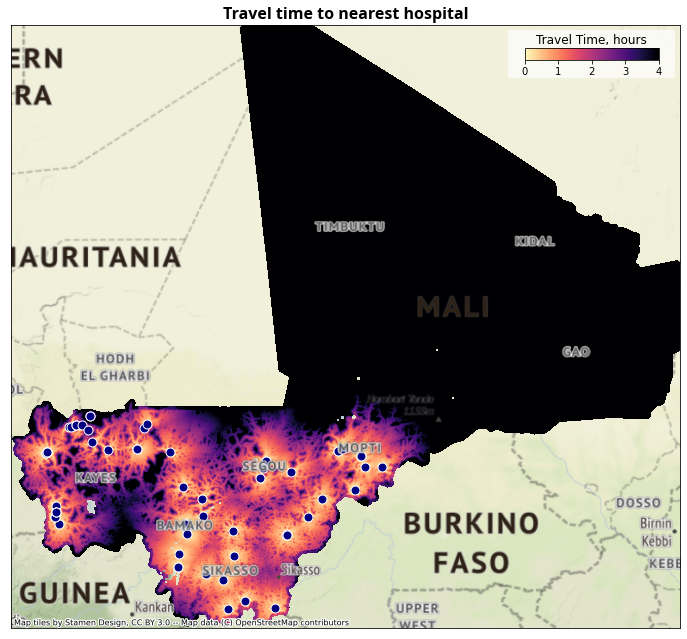
4.3. Summarize population within x hours#
Show code cell source
thresh = 2 # 2
pop_120_heatlh = pop*(tt_health<=thresh)
pop_120_hospital = pop*(tt_hospital<=thresh)
zs_pop = pd.DataFrame(zonal_stats(adm2, pop.filled(), affine=pop_surf.transform, stats='sum', nodata=pop_surf.nodata)).rename(columns={'sum':'pop'})
zs_lt_120_health = pd.DataFrame(zonal_stats(adm2, pop_120_heatlh.filled(), affine=pop_surf.transform, stats='sum', nodata=pop_surf.nodata)).rename(columns={'sum':'pop_120_health'})
zs_lt_120_hospital = pd.DataFrame(zonal_stats(adm2, pop_120_hospital.filled(), affine=pop_surf.transform, stats='sum', nodata=pop_surf.nodata)).rename(columns={'sum':'pop_120_hospital'})
zs = zs_pop.join(zs_lt_120_health).join(zs_lt_120_hospital)
zs.loc[:, "health_pct"] = zs.loc[:, "pop_120_health"]/zs.loc[:, "pop"]
zs.loc[:, "hospital_pct"] = zs.loc[:, "pop_120_hospital"]/zs.loc[:, "pop"]
res = adm2.join(zs)
Summary stats
Show code cell source
print(f"Summary of % of pop. within {thresh} hr. of health facility \n {res.health_pct.describe()}")
print("")
print(f"Summary of % of pop. within {thresh} hr. of hospital \n {res.hospital_pct.describe()}")
Summary of % of pop. within 2 hr. of health facility
count 55.000000
mean 0.887105
std 0.209835
min 0.165766
25% 0.922011
50% 0.984395
75% 0.998587
max 1.000000
Name: health_pct, dtype: float64
Summary of % of pop. within 2 hr. of hospital
count 55.000000
mean 0.612836
std 0.411968
min 0.000000
25% 0.035095
50% 0.823605
75% 0.951852
max 1.000000
Name: hospital_pct, dtype: float64
4.4. Access to roads#
Percentage of health facilities having direct access to an all-season road, by district (admin-2 level).
All-season road defined as primary, secondary or tertiary using the OpenStreetMap classification.
Direct access defined as being within 100 meters of a road.
4.4.1. Load OSM roads and define classification#
{
'motorway': 'OSMLR level 1',
'motorway_link': 'OSMLR level 1',
'trunk': 'OSMLR level 1',
'trunk_link': 'OSMLR level 1',
'primary': 'OSMLR level 1',
'primary_link': 'OSMLR level 1',
'secondary': 'OSMLR level 2',
'secondary_link': 'OSMLR level 2',
'tertiary': 'OSMLR level 2',
'tertiary_link': 'OSMLR level 2',
'unclassified': 'OSMLR level 3',
'unclassified_link': 'OSMLR level 3',
'residential': 'OSMLR level 3',
'residential_link': 'OSMLR level 3',
'track': 'OSMLR level 4',
'service': 'OSMLR level 4'
}
Show code cell source
roads = gpd.read_file(join(input_dir, 'osm', iso3, 'gis_osm_roads_free_1.shp'))
adm2_json = json.loads(adm2.to_json())
epsg = epsg_get(adm2_json)
roads = roads.to_crs(epsg)
roads['OSMLR'] = roads['fclass'].map(osm.OSMLR_Classes)
def get_num(x):
try:
return(int(x))
except:
return(5)
roads['OSMLR_num'] = roads['OSMLR'].apply(lambda x: get_num(str(x)[-1]))
## Calculate buffers
roads_100m = roads.copy()
roads_100m['geometry'] = roads_100m['geometry'].apply(lambda x: x.buffer(100))
roads_2km = roads.copy()
roads_2km['geometry'] = roads_2km['geometry'].apply(lambda x: x.buffer(2000))
health = health.to_crs(epsg)
# hospitals = hospitals.to_crs(epsg)
## Intersect roads and buffer
# roads_1_100m = roads_100m.loc[roads_100m.OSMLR_num<=1].unary_union
roads_2_100m = roads_100m.loc[roads_100m.OSMLR_num<=2].unary_union
# roads_3 = roads.loc[roads.OSMLR_num<=3].unary_union
# roads_4 = roads.loc[roads.OSMLR_num<=4].unary_union
# health.loc[:, "bool_1_100m"] = health.intersects(roads_1_100m)
health.loc[:, "bool_2_100m"] = health.intersects(roads_2_100m)
# health.loc[:, "bool_3"] = health.intersects(roads_3)
# health.loc[:, "bool_4"] = health.intersects(roads_4)
# roads_1_2km = roads_2km.loc[roads_2km.OSMLR_num<=1].unary_union
roads_2_2km = roads_2km.loc[roads_2km.OSMLR_num<=2].unary_union
# roads_3 = roads.loc[roads.OSMLR_num<=3].unary_union
# roads_4 = roads.loc[roads.OSMLR_num<=4].unary_union
# health.loc[:, "bool_1_2km"] = health.intersects(roads_1_2km)
health.loc[:, "bool_2_2km"] = health.intersects(roads_2_2km)
# health.loc[:, "bool_3"] = health.intersects(roads_3)
# health.loc[:, "bool_4"] = health.intersects(roads_4)
health = health.to_crs(adm2.crs)
facilities = gpd.sjoin(health, adm2[['OBJECTID', 'WB_ADM2_CO', 'geometry']], how='left')
## Get percentages
# res_osmlr = facilities[['bool_1_100m','bool_2_100m','bool_1_2km','bool_2_2km','WB_ADM2_CO']].groupby('WB_ADM2_CO').sum()
res_osmlr = facilities[['bool_2_100m','bool_2_2km','WB_ADM2_CO']].groupby('WB_ADM2_CO').sum()
res_count = facilities[['WB_ADM2_CO','bool_2_100m']].groupby('WB_ADM2_CO').count().rename(columns={'bool_2_100m':'count'})
res_osmlr_pct = res_osmlr.apply(lambda x: x/res_count['count'])
res = res.merge(res_osmlr_pct, left_on="WB_ADM2_CO", right_index=True)
4.4.2. Maps of Access to Roads#
Show code cell source
figsize = (12,12)
fig, ax = plt.subplots(1, 1, figsize = figsize)
fonttitle = {'fontname':'Open Sans','weight':'bold','size':16}
ax.set_title("Access to Roads - Direct Access", fontdict=fonttitle)
ax.get_xaxis().set_visible(False) # plt.axis('off')
ax.get_yaxis().set_visible(False)
res.plot(
ax=ax, column='bool_2_100m', cmap='Reds_r', legend=True,
scheme='user_defined', alpha=1, linewidth=0.2, edgecolor='black',
classification_kwds = {'bins': [0.2,0.4,0.6,0.8,1]},
legend_kwds = {
'title': "% of health facilities within \n 100m of an all-season road",
'fontsize': 10,
'fmt': "{:.0%}",
'title_fontsize': 12
}
)
ctx.add_basemap(ax, source=ctx.providers.Stamen.TerrainLabels, crs='EPSG:4326', zorder=1)
ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10, alpha=0.75)
plt.savefig(os.path.join(out_folder, "Roads_100m.png"), dpi=150, bbox_inches='tight', facecolor='white')
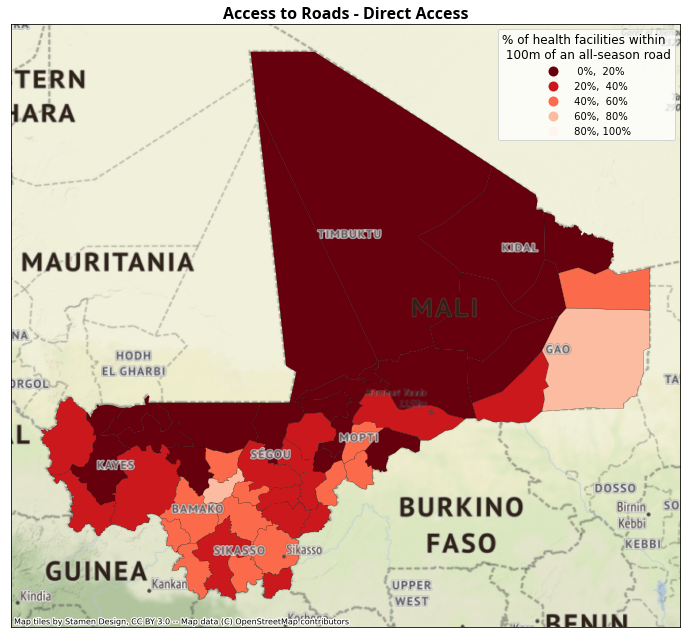
Show code cell source
figsize = (12,12)
fig, ax = plt.subplots(1, 1, figsize = figsize)
fonttitle = {'fontname':'Open Sans','weight':'bold','size':16}
ax.set_title("Access to Roads - 2 km", fontdict=fonttitle)
ax.get_xaxis().set_visible(False) # plt.axis('off')
ax.get_yaxis().set_visible(False)
res.plot(
ax=ax, column='bool_2_2km', cmap='Reds_r', legend=True, linewidth=0.2, edgecolor='black',
scheme='user_defined', alpha=1,
classification_kwds = {'bins': [0.2,0.4,0.6,0.8,1]},
legend_kwds = {
'title': "% of health facilities within \n 2km of an all-season road",
'fontsize': 10,
'fmt': "{:.0%}",
'title_fontsize': 12
}
)
ctx.add_basemap(ax, source=ctx.providers.Stamen.TerrainLabels, crs='EPSG:4326', zorder=1)
ctx.add_basemap(ax, source=ctx.providers.Stamen.Terrain, crs='EPSG:4326', zorder=-10, alpha=0.75)
plt.savefig(os.path.join(out_folder, "Roads_2km.png"), dpi=150, bbox_inches='tight', facecolor='white')
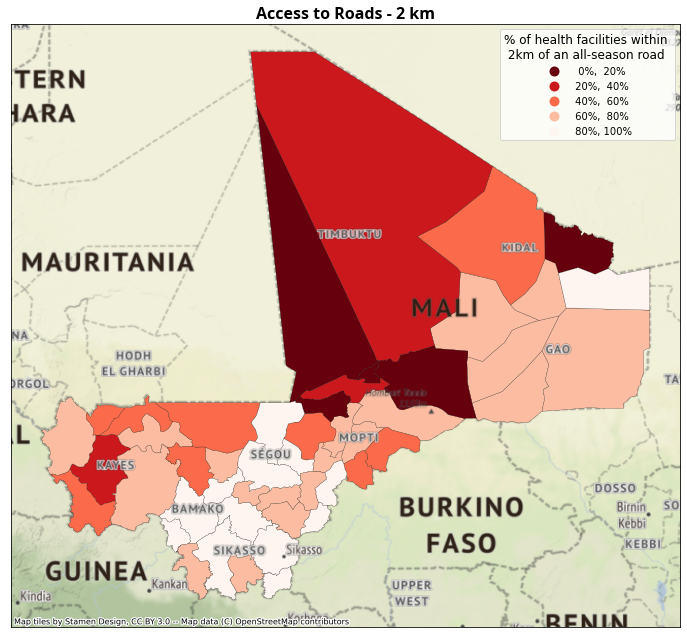
4.5. Access to health disaggregated by wealth quintile#
Categorize population grid by wealth quintiles, and then summarize the population with access to health (within 2 hours of health facility or hospital)
Show code cell source
fb = pd.read_csv(os.path.join(input_dir, 'facebook', f'{iso3.lower()}_relative_wealth_index.csv'))
fb_geoms = [Point(xy) for xy in zip(fb.longitude, fb.latitude)]
fb_geo = gpd.GeoDataFrame(fb, crs='EPSG:4326', geometry=fb_geoms)
### Summarize access with facebook grid
fb_zs = pd.DataFrame(zonal_stats(fb_geo, tt_health.filled(), affine=tt_health_rio.transform, stats='mean', nodata=tt_health_rio.nodata)).rename(columns={'mean':'tt_health'})
fb_zs_hosp = pd.DataFrame(zonal_stats(fb_geo, tt_hospital.filled(), affine=tt_hospital_rio.transform, stats='mean', nodata=tt_hospital_rio.nodata)).rename(columns={'mean':'tt_hospital'})
fb_geo = fb_geo.join(fb_zs).join(fb_zs_hosp)
fb_geo.loc[:, "rwi_cut"] = pd.qcut(fb_geo['rwi'], [0, .2, .4, .6, .8, 1.], labels=['lowest', 'second-lowest', 'middle', 'second-highest', 'highest'])
fb_geo = gpd.sjoin(fb_geo, adm2[['WB_ADM2_CO', 'WB_ADM2_NA', 'WB_ADM1_NA', 'geometry']])
### Merge population from Facebook
population = pd.read_csv(join(input_dir, 'facebook', f'{iso3.lower()}_general_2020.csv'))
population = population.rename(columns={f'{iso3.lower()}_general_2020': 'pop_2020'})
population['quadkey'] = population.apply(lambda x: str(quadkey.from_geo((x['latitude'], x['longitude']), 14)), axis=1)
bing_tile_z14_pop = population.groupby('quadkey', as_index=False)['pop_2020'].sum()
fb_geo['quadkey'] = fb_geo.apply(lambda x: str(quadkey.from_geo((x['latitude'], x['longitude']), 14)), axis=1)
rwi = fb_geo.merge(bing_tile_z14_pop[['quadkey', 'pop_2020']], on='quadkey', how='inner')
rwi.loc[:, "tt_health_bool"] = rwi.tt_health<=2
rwi.loc[:, "tt_hospital_bool"] = rwi.tt_hospital<=2
### Aggregate at country level
rwi_pop_adm0 = rwi[['rwi_cut', 'pop_2020']].groupby(['rwi_cut']).sum()
rwi_health_adm0 = rwi.loc[rwi.tt_health_bool==True, ['rwi_cut', 'pop_2020']].groupby(['rwi_cut']).sum().rename(columns={'pop_2020':'pop_120_health'})
rwi_hospital_adm0 = rwi.loc[rwi.tt_hospital_bool==True, ['rwi_cut', 'pop_2020']].groupby(['rwi_cut']).sum().rename(columns={'pop_2020':'pop_120_hospital'})
res_rwi_adm0 = rwi_pop_adm0.join(rwi_health_adm0).join(rwi_hospital_adm0)
res_rwi_adm0.loc[:, "health_pct"] = res_rwi_adm0['pop_120_health']/res_rwi_adm0['pop_2020']
res_rwi_adm0.loc[:, "hospital_pct"] = res_rwi_adm0['pop_120_hospital']/res_rwi_adm0['pop_2020']
res_rwi_adm0.reset_index(inplace=True)
res_rwi_adm0 = res_rwi_adm0[['rwi_cut', 'health_pct', 'hospital_pct']].melt(id_vars='rwi_cut', var_name="type", value_name='pct')
res_rwi_adm0.type = res_rwi_adm0.type.str.strip("_pct")
res_rwi_adm0.loc[res_rwi_adm0.type=="health", "type"] = 'health facility'
4.5.1. Chart access to health by wealth quintile#
Show code cell source
(
ggplot(res_rwi_adm0, aes(x="rwi_cut", y="pct", fill="type"))
+ geom_col(width=0.5, stat='identity', position='dodge')
+ labs(
x="", y="% of pop", title="Population Access to Health Facilities by Wealth Quintile",
fill="Within 2 hr. of"
)
+ theme_minimal()
+ scale_y_continuous(labels=percent_format())
# + theme(legend_position='bottom')
)
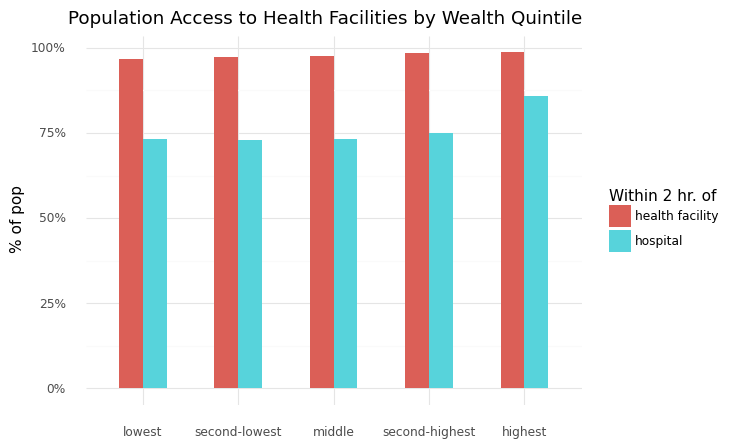
<ggplot: (8730507465969)>
4.5.2. Aggregate to province-level (admin 1)#
Show code cell source
rwi_pop = rwi[['WB_ADM1_NA', 'rwi_cut', 'pop_2020']].groupby(['WB_ADM1_NA', 'rwi_cut']).sum()
rwi_health = rwi.loc[rwi.tt_health_bool==True, ['WB_ADM1_NA', 'rwi_cut', 'pop_2020']].groupby(['WB_ADM1_NA', 'rwi_cut']).sum().rename(columns={'pop_2020':'pop_120_health'})
rwi_hospital = rwi.loc[rwi.tt_hospital_bool==True, ['WB_ADM1_NA', 'rwi_cut', 'pop_2020']].groupby(['WB_ADM1_NA', 'rwi_cut']).sum().rename(columns={'pop_2020':'pop_120_hospital'})
res_rwi = rwi_pop.join(rwi_health).join(rwi_hospital)
res_rwi.reset_index(inplace=True)
res_rwi.loc[:, "health_pct"] = res_rwi['pop_120_health']/res_rwi['pop_2020']
res_rwi.loc[:, "hospital_pct"] = res_rwi['pop_120_hospital']/res_rwi['pop_2020']
4.5.3. Chart access to health by wealth quintile, by province#
Show code cell source
(
ggplot(res_rwi, aes(x="rwi_cut", y="health_pct", fill="rwi_cut"))
+ geom_col(width=0.5)
+ labs(
x="", y="share of pop.", title="Population within 2hr. of a health facility",
fill="Wealth Quintile"
)
+ theme_minimal()
+ theme(axis_text_x=element_blank(), axis_ticks_major_x=element_blank())
+ facet_wrap("WB_ADM1_NA")
+ scale_y_continuous(labels=percent_format())
)
/home/wb514197/.conda/envs/graph/lib/python3.7/site-packages/plotnine/layer.py:381: PlotnineWarning: position_stack : Removed 4 rows containing missing values.

<ggplot: (8730379075285)>
Show code cell source
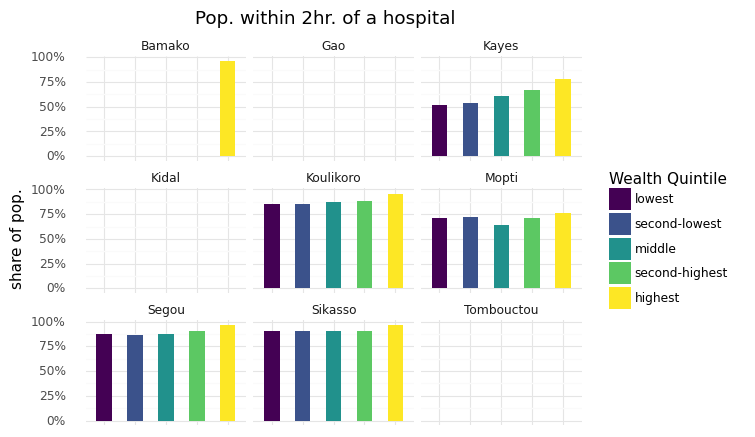
(
ggplot(res_rwi, aes(x="rwi_cut", y="hospital_pct", fill="rwi_cut"))
+ geom_col(width=0.5)
+ labs(
x="", y="share of pop.", title="Pop. within 2hr. of a hospital",
fill="Wealth Quintile"
)
+ theme_minimal()
+ theme(axis_text_x=element_blank(), axis_ticks_major_x=element_blank())
+ facet_wrap("WB_ADM1_NA")
+ scale_y_continuous(labels=percent_format())
)
/home/wb514197/.conda/envs/graph/lib/python3.7/site-packages/plotnine/layer.py:381: PlotnineWarning: position_stack : Removed 19 rows containing missing values.
<ggplot: (8730379028229)>