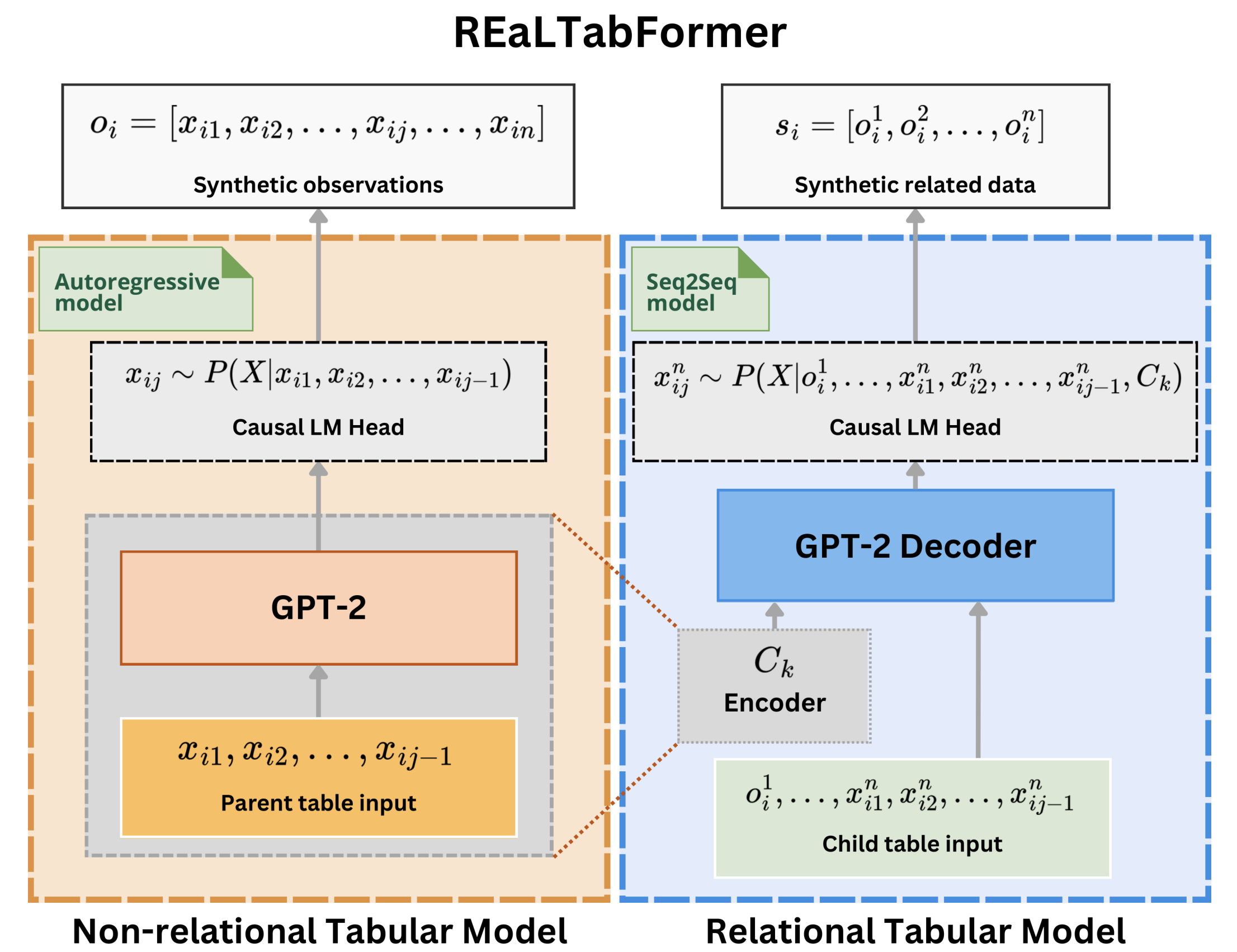
"""This module contains the implementation for the sampling
algorithms used for tabular and relational data generation.
"""
import logging
import warnings
from typing import Any, Dict, List, Optional, Union
import datasets
import numpy as np
import pandas as pd
import torch
from torch.utils.data import DataLoader
from tqdm.auto import tqdm
from transformers import DefaultDataCollator, PreTrainedModel
from .data_utils import (
INVALID_NUMS_RE,
NUMERIC_NA_TOKEN,
ModelType,
SpecialTokens,
decode_column_values,
decode_partition_numeric_col,
decode_processed_column,
fix_multi_decimal,
is_datetime_col,
is_numeric_col,
is_numeric_datetime_col,
make_dataset,
process_data,
)
from .rtf_exceptions import SampleEmptyError, SampleEmptyLimitError
from .rtf_validators import ObservationValidator
[docs]class REaLSampler:
def __init__(
self,
model_type: str,
model: PreTrainedModel,
vocab: Dict,
processed_columns: List,
max_length: int,
col_size: int,
col_idx_ids: Dict,
columns: List,
datetime_columns: List,
column_dtypes: Dict,
column_has_missing: Dict,
drop_na_cols: List,
col_transform_data: Dict,
random_state: Optional[int] = 1029,
device="cuda",
) -> None:
self.model_type = model_type
self.vocab = vocab
self.processed_columns = processed_columns
self.col_size = col_size # relational_col_size or tabular_col_size
self.col_idx_ids = col_idx_ids
self.max_length = max_length # relational_max_length or tabular_max_length
self.columns = columns
self.datetime_columns = datetime_columns
self.column_dtypes = column_dtypes
self.column_has_missing = column_has_missing
self.drop_na_cols = drop_na_cols
self.col_transform_data = col_transform_data
self.random_state = random_state
self.device = torch.device(device)
if model.device != self.device:
self.model = model.to(self.device)
else:
self.model = model
self.invalid_gen_samples = 0
self.total_gen_samples = 0
# Set the model to eval mode
self.model.eval()
[docs] def _prefix_allowed_tokens_fn(self, batch_id, input_ids) -> List:
# https://huggingface.co/docs/transformers/v4.24.0/en/main_classes/text_generation#transformers.generation_utils.GenerationMixin.generate.prefix_allowed_tokens_fn
raise NotImplementedError
[docs] def _convert_to_table(self, synth_df: pd.DataFrame) -> pd.DataFrame:
# Perform additional standardization
# processing.
synth_df = synth_df[sorted(synth_df.columns)]
synth_df.columns = synth_df.columns.map(decode_processed_column)
# Order based on the original data columns.
try:
synth_df = synth_df[self.columns]
except KeyError:
pass
for col in self.datetime_columns:
# Attempt to transform datetime data
# from the encoded timestamp into datetime.
try:
# Add the `mean_date` that we have subtracted during
# fitting of the data.
series = synth_df[col].copy()
series[series.notna()] += self.col_transform_data[col].get(
"mean_date", 0
)
# Multiply by 1e9 since we divided by this
# the original timestamp in the processing step.
synth_df[col] = pd.to_datetime(series * 1e9)
except Exception:
pass
# Attempt to cast the synthetic data to
# the original data types.
valid_idx = set(range(len(synth_df)))
_synth_df = []
for c, d in self.column_dtypes.items():
try:
series = synth_df[c]
if pd.api.types.is_numeric_dtype(d):
if self.column_has_missing[c]:
# If original data type is numeric,
# attempt to replace the <NA> token
# with NaN for casting to work.
series = series.replace("<NA>", "NaN")
if series.dtype == "object":
# Explicitly unify the data type
# in case we have mixed types in the series.
series = series.astype(str)
nan_idx = None
if self.column_has_missing[c]:
# Values where nan was generated first is treated
# as a valid nan value.
nan_idx = series.str.startswith(NUMERIC_NA_TOKEN)
series.loc[nan_idx] = "NaN"
# Removed indices of invalid values. Invalid values
# are those where nan was generated somewhere after
# the first token set for the variable or that any
# non-numeric value is present.
re_invalid_pattern = INVALID_NUMS_RE
# The initially changed "NaN" above will be
# included by the pattern. Make sure we add indices
# back to the valid_idx list.
invalid_idx = series.str.contains(
re_invalid_pattern, regex=True
)
valid_idx = valid_idx.difference(np.where(invalid_idx)[0])
if nan_idx is not None:
valid_idx = valid_idx.union(np.where(nan_idx)[0])
# Temporarily set them to NaN, but we will remove these
# observations later.
series.loc[invalid_idx] = "NaN"
elif pd.api.types.is_object_dtype(d):
# In case the values in this column is "integral"
# but are cast as object in the real data, let's try
# to convert them first to int before casting to the
# correct object type.
# Not doing this will incorrectly generate "float"-looking
# data for object types, but are integral, in the
# fb-comments dataset.
try:
series = series.astype(pd.Int64Dtype()).astype(str)
except TypeError:
pass
except ValueError:
pass
if series.dtype != d:
# To speed up a bit, only cast if the dtypes
# is not the same as the expected type.
series = series.astype(d)
_synth_df.append(series)
except Exception: # nolint
_synth_df.append(synth_df[c])
synth_df = pd.concat(_synth_df, axis=1)
# We expect columns that have no missing values in the
# training data should have no <NA> in them.
with_missing_cols = [
col for col, missing in self.column_has_missing.items() if missing
]
if with_missing_cols:
synth_df[with_missing_cols] = (
synth_df[with_missing_cols]
.replace(f".*{NUMERIC_NA_TOKEN}+.*", "NaN", regex=True)
.replace("<NA>", "NaN")
.replace("NaN", np.nan)
)
if len(valid_idx) > 0:
synth_df = synth_df.iloc[sorted(valid_idx)]
return synth_df
[docs] def _generate(
self,
device: torch.device,
as_numpy: Optional[bool] = True,
constrain_tokens_gen: Optional[bool] = True,
**generate_kwargs,
) -> Union[torch.tensor, np.ndarray]:
# This leverages the generic interface of HuggingFace transformer models' `.generate` method.
# Refer to the transformers documentation for valid arguments to `generate_kwargs`.
self.model.eval()
if constrain_tokens_gen:
generate_kwargs["prefix_allowed_tokens_fn"] = self._prefix_allowed_tokens_fn
vocab = (
self.vocab
if self.model_type == ModelType.tabular
else self.vocab["decoder"]
)
# Make sure that the [RMASK] token will never be generated.
RMASK_ID = vocab["token2id"][SpecialTokens.RMASK]
if generate_kwargs["suppress_tokens"] is None:
generate_kwargs["suppress_tokens"] = [RMASK_ID]
else:
generate_kwargs["suppress_tokens"].append(RMASK_ID)
if "bos_token_id" not in generate_kwargs:
generate_kwargs["bos_token_id"] = vocab["token2id"][SpecialTokens.BOS]
if "pad_token_id" not in generate_kwargs:
generate_kwargs["pad_token_id"] = vocab["token2id"][SpecialTokens.PAD]
if "eos_token_id" not in generate_kwargs:
generate_kwargs["eos_token_id"] = vocab["token2id"][SpecialTokens.EOS]
_samples = self.model.generate(**generate_kwargs)
if as_numpy:
if device == torch.device("cpu"):
_samples = _samples.numpy()
else:
_samples = _samples.cpu().numpy()
return _samples
[docs] def _validate_synth_sample(self, synth_sample: pd.DataFrame) -> pd.DataFrame:
# Validate data
valid_mask = []
# Validate that the generated value for a column is correct.
# Use the column identifier as basis for the validation.
# Is this useful when we actually filter the vocabulary
# during generation???
# Let's try removing this for now and see what happens... XD
# We remove this because the operations here are expensive, and
# slow down the sampling.
for col in synth_sample.columns:
valid_mask.append(synth_sample[col].str.startswith(col))
valid_mask = pd.concat(valid_mask, axis=1).all(axis=1)
synth_sample = synth_sample.loc[valid_mask]
if synth_sample.empty:
# Handle this exception in the sampling function.
raise SampleEmptyError(in_size=len(valid_mask))
return synth_sample
[docs] def _recover_data_values(self, synth_sample: pd.DataFrame) -> pd.DataFrame:
processed_columns = pd.Index(self.processed_columns)
numeric_datetime_cols = processed_columns[
processed_columns.map(is_numeric_datetime_col)
]
_tmp_synth_df = []
_tmp_synth_cols = []
# Get the actual column name for numerically
# transformed data.
numeric_col_group = numeric_datetime_cols.groupby(
numeric_datetime_cols.map(decode_partition_numeric_col)
)
for col, c_group in numeric_col_group.items():
# Aggregate the partitioned data for the actual column.
group_series = synth_sample[c_group].sum(axis=1)
if group_series.dtype == "object":
# This automatically casts into numeric type if all values are
# valid numbers.
group_series = group_series.str.lstrip("0")
# If all values are zeros for a group, then it will be left
# empty. Let's explicitly set the zero value back.
# TODO: review other use cases that this may not be correct.
# For example, zero may not be in the domain of the
# variable `CompetitionDistance` in the Rossmann dataset.
group_series[group_series == ""] = 0
# If the data is still an object type, try to fix potential
# errors.
if group_series.dtype == "object":
if is_numeric_col(col):
try:
# This usually happens when the decimal point was generated
# multiple times in the value. This simply removes the succeeding
# occurence of the decimal point.
group_series = (
group_series.apply(fix_multi_decimal)
.astype(float)
.fillna(pd.NA)
)
except Exception:
pass
elif is_datetime_col(col):
# We expect that timestamp data is represented fully
# by numbers. Just remove all non-numeric characters.
# This may introduce invalid values somehow, but validators
# can later be implemented to remove these.
group_series = (
group_series.str.replace("[^0-9]", "", regex=True)
.map(lambda x: int(x) if x == x else None)
.fillna(pd.NA)
)
else:
raise ValueError(f"Unknown column dtype for {col}...")
if is_numeric_col(col):
try:
# Try to force convert values to Int64Dtype
group_series = group_series.astype(pd.Int64Dtype())
except TypeError:
pass
except ValueError:
# Example:
# ValueError: invalid literal for int() with base 10: '1271.0942'
pass
_tmp_synth_df.append(group_series)
_tmp_synth_cols.append(col)
# Add data for categorical columns
for col in synth_sample.columns:
if col in numeric_datetime_cols:
continue
_tmp_synth_df.append(synth_sample[col])
_tmp_synth_cols.append(col)
synth_df: pd.DataFrame = pd.concat(_tmp_synth_df, axis=1).reset_index(
drop="index"
)
synth_df.columns = _tmp_synth_cols
synth_df.index = synth_sample.index
return synth_df
[docs] def _processes_sample(
self,
sample_outputs: np.ndarray,
vocab: Dict,
relate_ids: Optional[List[Any]] = None,
validator: Optional[ObservationValidator] = None,
) -> pd.DataFrame:
assert isinstance(sample_outputs, np.ndarray)
def _decode_tokens(s):
# No need to remove [BOS] and [EOS] tokens
# here, it will be handled later.
return [vocab["id2token"][i] for i in s]
if self.model_type == ModelType.tabular:
# Slice to remove the [BOS] and [EOS] tokens
synth_sample = pd.DataFrame(
[_decode_tokens(s)[1:-1] for s in sample_outputs],
columns=self.processed_columns,
)
else:
assert relate_ids is not None
_samples = [_decode_tokens(s) for s in sample_outputs]
# Unpack the tokens and remove any special tokens.
# Also perform segmentation of observations for
# the relational model.
group_ids = []
samples = []
for ix, (rel_id, dg) in enumerate(zip(relate_ids, _samples)):
group = []
ind: List[str] = []
for token_ix, v in enumerate(dg):
if v in [SpecialTokens.BMEM, SpecialTokens.EOS]:
if len(ind) > 0:
# Review later whether we should void
# the entire generation process for this
# input or not. For now, let's just
# throw this particular observation that didn't
# satisfy the expected col_size.
if len(ind) == self.col_size:
group.append(ind)
else:
logging.warning(
f"Discarding this observation for input index:{ix} with an invalid number of columns: {len(ind)}."
)
ind = []
if (v == SpecialTokens.EOS) and (token_ix > 0):
break
elif v in [SpecialTokens.BOS, SpecialTokens.EMEM]:
continue
elif v == SpecialTokens.PAD:
# This should not go here, but putting this just in case.
break
else:
ind.append(v)
group_ids.extend([rel_id] * len(group))
samples.extend(group)
# Create a unique index for observations
# that are supposed to be generated by the
# same input data.
synth_sample = pd.DataFrame(
samples, columns=self.processed_columns, index=group_ids
)
# Initial check for an empty sample frame.
if synth_sample.empty:
# Handle this exception in the sampling function.
raise SampleEmptyError(in_size=len(sample_outputs))
# # Is this useful when we actually filter the vocabulary
# # during generation???
# # Let's try removing this for now and see what happens... XD
# # We remove this because the operations here are expensive, and
# # slow down the sampling.
# synth_sample = self._validate_synth_sample(synth_sample)
# Extract the values
for col in synth_sample.columns:
# Filter only columns that we have explicitly processed.
# Since the column values have been previously validated
# to only contain values that match the column, then it
# is safe to just check the first row for this contraint.
if synth_sample[col].iloc[0].startswith(col):
synth_sample[col] = decode_column_values(synth_sample[col])
synth_df = self._recover_data_values(synth_sample)
logging.info(f"Generation stats: {synth_df.shape[0]}")
synth_df = self._convert_to_table(synth_df)
synth_df = self._validate_missing(synth_df)
synth_df = self._validate_data(synth_df, validator)
if synth_df.empty:
# Handle this exception in the sampling function.
raise SampleEmptyError(in_size=len(sample_outputs))
return synth_df
[docs] def _validate_data(
self, synth_df: pd.DataFrame, validator: Optional[ObservationValidator] = None
) -> pd.DataFrame:
if validator is not None:
synth_df = synth_df.loc[validator.validate_df(synth_df)]
return synth_df
[docs] def _validate_missing(self, synth_df: pd.DataFrame) -> pd.DataFrame:
# Drop the rows where any one of the columns that should not have
# a missing value have at least one.
return synth_df.dropna(subset=self.drop_na_cols)
[docs]class TabularSampler(REaLSampler):
"""Sampler class for tabular data generation."""
def __init__(
self,
model_type: str,
model: PreTrainedModel,
vocab: Dict,
processed_columns: List,
max_length: int,
col_size: int,
col_idx_ids: Dict,
columns: List,
datetime_columns: List,
column_dtypes: Dict,
column_has_missing: Dict,
drop_na_cols: List,
col_transform_data: Dict,
random_state: Optional[int] = 1029,
device="cuda",
) -> None:
super().__init__(
model_type,
model,
vocab,
processed_columns,
max_length,
col_size,
col_idx_ids,
columns,
datetime_columns,
column_dtypes,
column_has_missing,
drop_na_cols,
col_transform_data,
random_state,
device,
)
self.output_vocab = self.vocab
@staticmethod
[docs] def sampler_from_model(rtf_model, device: str = "cuda"):
device = torch.device(device)
assert rtf_model.tabular_max_length is not None
assert rtf_model.tabular_col_size is not None
assert rtf_model.col_transform_data is not None
return TabularSampler(
model_type=rtf_model.model_type,
model=rtf_model.model,
vocab=rtf_model.vocab,
processed_columns=rtf_model.processed_columns,
max_length=rtf_model.tabular_max_length,
col_size=rtf_model.tabular_col_size,
col_idx_ids=rtf_model.col_idx_ids,
columns=rtf_model.columns,
datetime_columns=rtf_model.datetime_columns,
column_dtypes=rtf_model.column_dtypes,
column_has_missing=rtf_model.column_has_missing,
drop_na_cols=rtf_model.drop_na_cols,
col_transform_data=rtf_model.col_transform_data,
random_state=rtf_model.random_state,
device=device,
)
[docs] def _prefix_allowed_tokens_fn(self, batch_id, input_ids) -> List:
# https://huggingface.co/docs/transformers/v4.24.0/en/main_classes/text_generation#transformers.generation_utils.GenerationMixin.generate.prefix_allowed_tokens_fn
# For the tabular data, len(input_ids) == 1 -> [BOS]
# Subtract by 1 since the first valid token has index zero in
# col_idx_ids while the input_ids already contains the [BOS] token.
return self.col_idx_ids.get(
len(input_ids) - 1, [self.vocab["token2id"][SpecialTokens.EOS]]
)
[docs] def sample_tabular(
self,
n_samples: int,
gen_batch: Optional[int] = 128,
device: Optional[str] = "cuda",
seed_input: Optional[Union[pd.DataFrame, Dict[str, Any]]] = None,
constrain_tokens_gen: Optional[bool] = True,
validator: Optional[ObservationValidator] = None,
continuous_empty_limit: int = 10,
suppress_tokens: Optional[List[int]] = None,
forced_decoder_ids: Optional[List[List[int]]] = None,
**generate_kwargs,
) -> pd.DataFrame:
device = torch.device(device)
if self.model.device != device:
self.model = self.model.to(device)
self.model.eval()
synth_df = []
if seed_input is None:
generated = torch.tensor(
[self.vocab["token2id"][SpecialTokens.BOS] for _ in range(1)]
).unsqueeze(0)
else:
generated = self._process_seed_input(seed_input=seed_input)
generated = generated.to(self.model.device)
with tqdm(total=n_samples) as pbar:
pbar_num_gen = 0
num_generated = 0
empty_limit = continuous_empty_limit
while num_generated < n_samples:
# https://huggingface.co/docs/transformers/internal/generation_utils
sample_outputs = self._generate(
device=device,
as_numpy=True,
constrain_tokens_gen=constrain_tokens_gen,
inputs=generated,
do_sample=True,
max_length=self.max_length,
num_return_sequences=gen_batch,
bos_token_id=self.vocab["token2id"][SpecialTokens.BOS],
pad_token_id=self.vocab["token2id"][SpecialTokens.PAD],
eos_token_id=self.vocab["token2id"][SpecialTokens.EOS],
suppress_tokens=suppress_tokens,
forced_decoder_ids=forced_decoder_ids,
**generate_kwargs,
)
self.total_gen_samples += len(sample_outputs)
self.invalid_gen_samples += len(sample_outputs)
try:
synth_sample = self._processes_sample(
sample_outputs=sample_outputs,
vocab=self.vocab,
validator=validator,
)
empty_limit = continuous_empty_limit
self.invalid_gen_samples -= len(synth_sample)
except SampleEmptyError as exc:
logging.warning("This batch returned an empty valid synth_sample!")
empty_limit -= 1
if empty_limit <= 0:
raise SampleEmptyLimitError(
f"The model has generated empty sample batches for {continuous_empty_limit} consecutive rounds!"
) from exc
continue
num_generated += len(synth_sample)
synth_df.append(synth_sample)
# Update process bar
pbar.update(num_generated - pbar_num_gen)
pbar_num_gen = num_generated
synth_df = pd.concat(synth_df).sample(
n=n_samples, replace=False, random_state=self.random_state
)
synth_df = synth_df.reset_index(drop="index")
print(
f"Generated {self.invalid_gen_samples} invalid samples out of total {self.total_gen_samples} samples generated. Sampling efficiency is: {100 * (1 - self.invalid_gen_samples / self.total_gen_samples):.4f}%"
)
return synth_df
[docs] def predict(
self,
data: pd.DataFrame,
target_col: str,
target_pos_val: Any = None,
batch: int = 32,
obs_sample: int = 30,
fillunk: bool = True,
device: str = "cuda",
disable_progress_bar: bool = True,
**generate_kwargs,
) -> pd.Series:
"""
fillunk: Fill unknown tokens with the mode of the batch.
target_pos_val: Categorical value for the positive target. This is produces a
one-to-many prediction relative to `target_pos_val` for targets that are multi-categorical.
"""
device = torch.device(device)
if self.model.device != device:
self.model = self.model.to(device)
self.model.eval()
preds = []
unk_id = self.vocab["token2id"][SpecialTokens.UNK]
if target_col and target_col in data.columns:
data = data.drop(target_col, axis=1)
if disable_progress_bar:
datasets.utils.disable_progress_bar()
for i in range(0, len(data), batch):
seed_data = self._process_seed_input(data.iloc[i : i + batch])
if fillunk:
mode = seed_data.mode(dim=0).values
seed_data[seed_data == unk_id] = torch.tile(mode, (len(seed_data), 1))[
seed_data == unk_id
]
sample_outputs = self._generate(
device=device,
do_sample=True,
num_return_sequences=obs_sample,
input_ids=seed_data.to(device),
max_length=self.max_length,
suppress_tokens=[unk_id],
**generate_kwargs,
)
synth_sample = self._processes_sample(
sample_outputs=sample_outputs,
vocab=self.vocab,
validator=None,
)
# Reset the index so that we are sure that
# the index is monotonically increasing.
# There could be instances where some generation
# problems arise for some records, we don't
# handle that here for now.
synth_sample.reset_index(drop=True, inplace=True)
preds.extend(
synth_sample.groupby(synth_sample.index // obs_sample)[
target_col
].apply(
lambda x: (target_pos_val == x).mean()
if target_pos_val is not None
else x.mean()
)
)
if disable_progress_bar:
datasets.utils.enable_progress_bar()
return pd.Series(preds, index=data.index)
[docs]class RelationalSampler(REaLSampler):
"""Sampler class for relational data generation."""
def __init__(
self,
model_type: str,
model: PreTrainedModel,
vocab: Dict,
processed_columns: List,
max_length: int,
col_size: int,
col_idx_ids: Dict,
columns: List,
datetime_columns: List,
column_dtypes: Dict,
column_has_missing: Dict,
drop_na_cols: List,
col_transform_data: Dict,
in_col_transform_data: Dict,
random_state: Optional[int] = 1029,
device="cuda",
) -> None:
super().__init__(
model_type,
model,
vocab,
processed_columns,
max_length,
col_size,
col_idx_ids,
columns,
datetime_columns,
column_dtypes,
column_has_missing,
drop_na_cols,
col_transform_data,
random_state,
device,
)
self.output_vocab = self.vocab["decoder"]
self.in_col_transform_data = in_col_transform_data
@staticmethod
[docs] def sampler_from_model(rtf_model, device: str = "cuda"):
device = torch.device(device)
assert rtf_model.relational_max_length is not None
assert rtf_model.relational_col_size is not None
assert rtf_model.col_transform_data is not None
assert rtf_model.in_col_transform_data is not None
return RelationalSampler(
model_type=rtf_model.model_type,
model=rtf_model.model,
vocab=rtf_model.vocab,
processed_columns=rtf_model.processed_columns,
max_length=rtf_model.relational_max_length,
col_size=rtf_model.relational_col_size,
col_idx_ids=rtf_model.col_idx_ids,
columns=rtf_model.columns,
datetime_columns=rtf_model.datetime_columns,
column_dtypes=rtf_model.column_dtypes,
column_has_missing=rtf_model.column_has_missing,
drop_na_cols=rtf_model.drop_na_cols,
col_transform_data=rtf_model.col_transform_data,
in_col_transform_data=rtf_model.in_col_transform_data,
random_state=rtf_model.random_state,
device=device,
)
[docs] def sample_relational(
self,
input_unique_ids: Union[pd.Series, List],
input_df: Optional[pd.DataFrame] = None,
input_ids: Optional[torch.tensor] = None,
gen_batch: Optional[int] = 128,
device: Optional[str] = "cuda",
constrain_tokens_gen: Optional[bool] = True,
validator: Optional[ObservationValidator] = None,
continuous_empty_limit: Optional[int] = 10,
suppress_tokens: Optional[List[int]] = None,
forced_decoder_ids: Optional[List[List[int]]] = None,
related_num: Optional[Union[int, List[int]]] = None,
**generate_kwargs,
) -> pd.DataFrame:
# input_unique_ids: Corresponds to the unique identifier
# that will be used to link the input
# data to the generated values.
device = torch.device(device)
if self.model.device != device:
self.model = self.model.to(device)
self.model.eval()
input_unique_ids = list(input_unique_ids)
if input_ids is not None:
if isinstance(related_num, str):
warnings.warn(
f"The input provided is ids so related_num={related_num} is ignored."
)
related_num = None
generate_kwargs.update(self._get_min_max_length(related_num))
assert len(input_unique_ids) == len(input_ids)
input_ids = input_ids.to(device)
samples = self._generate(
device=device,
as_numpy=True,
constrain_tokens_gen=constrain_tokens_gen,
inputs=input_ids,
# num_return_sequences=gen_batch,
do_sample=True,
forced_decoder_ids=forced_decoder_ids,
suppress_tokens=suppress_tokens,
**generate_kwargs,
)
elif input_df is not None:
assert len(input_unique_ids) == input_df.shape[0]
# Create a fixed-size matrix to store the data filled with [PAD] token ids.
samples = np.ones((input_df.shape[0], self.max_length))
samples *= self.vocab["decoder"]["token2id"][SpecialTokens.PAD]
start = 0
if isinstance(related_num, str) and related_num in input_df.columns:
init_min_max_length = self._get_min_max_length(
input_df[related_num].max()
)
if init_min_max_length["max_length"] > samples.shape[1]:
samples = np.ones(
(input_df.shape[0], init_min_max_length["max_length"])
)
samples *= self.vocab["decoder"]["token2id"][SpecialTokens.PAD]
_input_unique_ids = []
input_df = input_df.copy()
# Make sure that we couple the `input_unique_ids` with
# its intended data when sorting and grouping.
input_df[NQ_COL] = input_unique_ids
input_df.sort_values(related_num, ascending=True, inplace=True)
for related_num, _input_df in input_df.groupby(related_num):
_input_unique_ids.append(_input_df.pop(NQ_COL))
generate_kwargs.update(self._get_min_max_length(related_num))
for _samples in self._sample_input_batch(
input_df=_input_df,
gen_batch=gen_batch,
device=device,
constrain_tokens_gen=constrain_tokens_gen,
suppress_tokens=suppress_tokens,
forced_decoder_ids=forced_decoder_ids,
**generate_kwargs,
):
end = start + len(_samples)
samples[start:end, : _samples.shape[1]] = _samples
start = end
input_unique_ids = pd.concat(_input_unique_ids)
else:
generate_kwargs.update(self._get_min_max_length(related_num))
if generate_kwargs["max_length"] > samples.shape[1]:
# Create a fixed-size matrix to store the data filled with [PAD] token ids.
samples = np.ones(
(input_df.shape[0], generate_kwargs["max_length"])
)
samples *= self.vocab["decoder"]["token2id"][SpecialTokens.PAD]
for _samples in self._sample_input_batch(
input_df=input_df,
gen_batch=gen_batch,
device=device,
constrain_tokens_gen=constrain_tokens_gen,
suppress_tokens=suppress_tokens,
forced_decoder_ids=forced_decoder_ids,
**generate_kwargs,
):
end = start + len(_samples)
samples[start:end, : _samples.shape[1]] = _samples
start = end
else:
raise ValueError("Either `input_ids` or `input_df` must not be None.")
synth_df = self._processes_sample(
sample_outputs=samples,
vocab=self.vocab["decoder"],
relate_ids=input_unique_ids,
validator=validator,
)
return synth_df
[docs] def _get_min_max_length(self, related_num):
# The `min_length = 2` corresponds to the ([EOS], [BOS])
# sequence that is used in the encoder-decoder
# model. This is why a related num of zero will have a
# max_length of (2 + 1) because we expect the next token
# in this sequence should be [EOS]. In the case where
# the `related_num > 0` we add `2` to col_size to account
# for the [BMEM] and the [EMEM] tokens.
min_length = 2
max_length = self.max_length
if related_num is not None:
if related_num >= 0:
min_length = min_length + ((self.col_size + 2) * related_num)
max_length = min_length + 1
else:
raise ValueError(
"The `related_num` must be greater than or equal to zero."
)
return dict(min_length=min_length, max_length=max_length)
[docs] def _get_relational_col_idx_ids(self, len_ids: int) -> List:
"""This method returns the true index given the generation step `i`.
col_size: The expected number of variables for a single observation.
This is equal to the number of columns.
### Generating constrained tokens per step
```
1 -> BOS
2 -> BMEM or EOS
3 -> col 0
...
3 + col_size -> col col_size - 1
3 + col_size + 1 -> EMEM
3 + col_size + 2 -> BMEM or EOS
3 + col_size + 3 -> col 0
```
"""
if len_ids == 0:
return_ids = [self.vocab["decoder"]["token2id"][SpecialTokens.EOS]]
elif len_ids == 1:
# This is the decoder_start_token_id and we should generate the [BOS] token next.
return_ids = [self.vocab["decoder"]["token2id"][SpecialTokens.BOS]]
else:
# Adjust such that idx = 0 produces either the [BMEM] or the [EOS] tokens.
idx = len_ids - 2
# Number of columns in the data for each observation
# self.col_size = self.relational_col_size
# Number of pads between observations [EMEM], ([BMEM] | [EOS])
num_pads = 2
# Derive `col_idx` such that at idx == 0, it will return -1 which
# maps to [[BMEM] or [EOS]]. Then idx == 1 will generate the first
# valid token, ..., then at idx == col_size, will generate the
# last valid token. At idx == col_size + 1, it will generate col_size
# which should generate the [[EMEM]] token.
col_idx = (idx % (self.col_size + num_pads)) - 1
assert -1 <= col_idx <= self.col_size
if col_idx < 0:
col_idx = -1
elif col_idx >= self.col_size:
col_idx = -2
return_ids = self.col_idx_ids[col_idx]
return return_ids
[docs] def _prefix_allowed_tokens_fn(self, batch_id, input_ids) -> List:
# https://huggingface.co/docs/transformers/v4.24.0/en/main_classes/text_generation#transformers.generation_utils.GenerationMixin.generate.prefix_allowed_tokens_fn
# For the relational data, len(input_ids) == 2 -> [EOS, BOS]
return self._get_relational_col_idx_ids(len(input_ids))