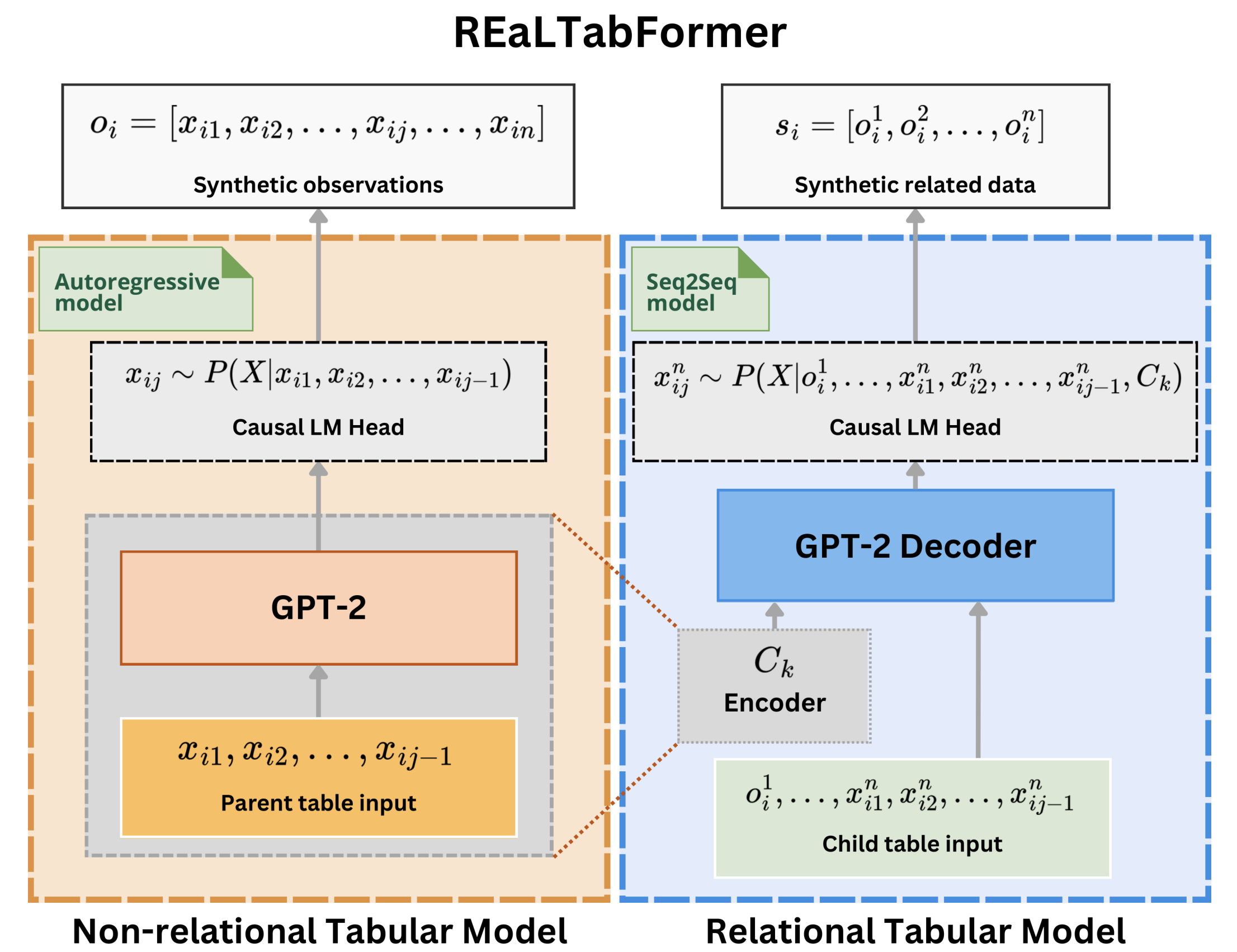
Source code for realtabformer.realtabformer
"""The REaLTabFormer implements the model training and data processing
for tabular and relational data.
"""
import json
import logging
import math
import os
import random
import shutil
import time
import warnings
from collections import OrderedDict
from pathlib import Path
from typing import Any, Dict, List, Optional, Union
import numpy as np
import pandas as pd
import torch
from datasets import Dataset
from sklearn.metrics.pairwise import manhattan_distances
# from sklearn.metrics import accuracy_score
from transformers import (
EarlyStoppingCallback,
EncoderDecoderConfig,
EncoderDecoderModel,
PreTrainedModel,
Seq2SeqTrainer,
Seq2SeqTrainingArguments,
Trainer,
TrainingArguments,
)
from transformers.models.gpt2 import GPT2Config, GPT2LMHeadModel
import realtabformer
from .data_utils import (
ModelFileName,
ModelType,
SpecialTokens,
TabularArtefact,
build_vocab,
make_dataset,
make_relational_dataset,
process_data,
)
from .rtf_analyze import SyntheticDataBench
from .rtf_datacollator import RelationalDataCollator
from .rtf_exceptions import SampleEmptyLimitError
from .rtf_sampler import RelationalSampler, TabularSampler
from .rtf_trainer import ResumableTrainer
from .rtf_validators import ObservationValidator
[docs]def _normalize_gpt2_state_dict(state_dict):
state = []
for key, value in state_dict.items():
if key.startswith("transformer."):
# The saved state prefixes the weight names
# with `transformer.` whereas the
# encoder expects the weight names to not
# have the prefix.
key = key.replace("transformer.", "")
state.append((key, value))
return OrderedDict(state)
[docs]def _validate_get_device(device: str) -> str:
if (device == "cuda") and (torch.cuda.device_count() == 0):
if torch.backends.mps.is_available():
_device = "mps"
else:
_device = "cpu"
warnings.warn(
f"The device={device} is not available, using device={_device} instead."
)
device = _device
return device
[docs]class REaLTabFormer:
def __init__(
self,
model_type: str,
tabular_config: Optional[GPT2Config] = None,
relational_config: Optional[EncoderDecoderConfig] = None,
parent_realtabformer_path: Optional[Path] = None,
freeze_parent_model: Optional[bool] = True,
checkpoints_dir: str = "rtf_checkpoints",
samples_save_dir: str = "rtf_samples",
epochs: int = 1000,
batch_size: int = 8,
random_state: int = 1029,
train_size: float = 1,
output_max_length: int = 512,
early_stopping_patience: int = 5,
early_stopping_threshold: float = 0,
mask_rate: float = 0,
numeric_nparts: int = 1,
numeric_precision: int = 4,
numeric_max_len: int = 10,
**training_args_kwargs,
) -> None:
"""Set up a REaLTabFormer instance.
Args:
model_type: Explicit declaration of which type of model will be used.
Can take `tabular` and `relational` as valid values.
tabular_config: GPT2Config instance to customize the GPT2 model for tabular data
generation.
relational_config: EncoderDecoderConfig instance that defines the encoder and decoder
configs for the encoder-decoder model used for the relational data generation. See
link for example: https://huggingface.co/docs/transformers/model_doc/encoder-decoder
parent_realtabformer_path: Path to a saved tabular REaLTabFormer model trained on the
parent table of a relational tabular data.
freeze_parent_model: Boolean flag indicating whether the parent-based encoder will be
frozen or not.
checkpoints_dir: Directory where the training checkpoints will be saved
samples_save_dir: Save the samples generated by this model in this directory.
epochs: Number of epochs for training the GPT2LM model. Use a large number of epochs to take advantage of the framework's optimal termination feature for the non-relational tabular data model. Defaults to 1000.
batch_size: Batch size used for training. Must be adjusted based on the available
compute resource. TrainingArguments is set to use `gradient_accumulation_steps=4`
which will have an effective batch_size of 32 for the default value.
train_size: Fraction of the data that will be passed to the `.fit` method that will
be used for training. The remaining will be used as validation data.
output_max_length: Truncation length for the number of output token ids in the
relational model. This limit applies to the processed data and not the raw number
of variables. This is not used in the tabular data model.
early_stopping_patience: Number of evaluation rounds without improvement before
stopping the training.
early_stopping_threshold: See link
https://huggingface.co/docs/transformers/main_classes/callback#transformers.EarlyStoppingCallback.early_stopping_threshold(float,
mask_rate: The rate of tokens in the transformed observation that will be replaced
with the [RMASK] token for regularization during training.
training_args_kwargs: Keyword arguments for the `TrainingArguments` used in training
the model. Arguments such as `output_dir`, `num_train_epochs`,
`per_device_train_batch_size`, `per_device_eval_batch_size` if passed will be
replaced by `checkpoints_dir`, `epochs`, `batch_size arguments`. The comprehensive
set of options can be found in
https://huggingface.co/docs/transformers/main/en/main_classes/trainer#transformers.TrainingArguments
"""
self.model: PreTrainedModel = None
# This will be set during and will also be deleted after training.
self.dataset = None
if model_type not in ModelType.types():
self._invalid_model_type(model_type)
self.model_type = model_type
if self.model_type == ModelType.tabular:
self._init_tabular(tabular_config)
elif self.model_type == ModelType.relational:
self.parent_vocab = None
self.parent_gpt2_config = None
self.parent_gpt2_state_dict = None
self.parent_col_transform_data = None
self.freeze_parent_model = freeze_parent_model
if parent_realtabformer_path is not None:
parent_realtabformer_path = Path(parent_realtabformer_path)
parent_config = json.loads(
(
parent_realtabformer_path / ModelFileName.rtf_config_json
).read_text()
)
self.parent_col_transform_data = parent_config["col_transform_data"]
self.parent_vocab = parent_config["vocab"]
self.parent_gpt2_config = parent_config["tabular_config"]
self.parent_gpt2_state_dict = _normalize_gpt2_state_dict(
torch.load(parent_realtabformer_path / ModelFileName.rtf_model_pt)
)
if output_max_length is None:
warnings.warn(
"The `output_max_length` is None. This could result to extended model training if the output length has large variations."
)
self.output_max_length = output_max_length
self._init_relational(relational_config)
else:
self._invalid_model_type(self.model_type)
self.checkpoints_dir = Path(checkpoints_dir)
self.samples_save_dir = Path(samples_save_dir)
self.epochs = epochs
self.batch_size = batch_size
self.early_stopping_patience = early_stopping_patience
self.early_stopping_threshold = early_stopping_threshold
self.training_args_kwargs = dict(
eval_strategy="steps",
output_dir=self.checkpoints_dir.as_posix(),
metric_for_best_model="loss", # This will be replaced with "eval_loss" if `train_size` < 1
overwrite_output_dir=True,
num_train_epochs=self.epochs,
per_device_train_batch_size=self.batch_size,
per_device_eval_batch_size=self.batch_size,
gradient_accumulation_steps=4,
remove_unused_columns=True,
logging_steps=100,
save_steps=100,
eval_steps=100,
load_best_model_at_end=True,
save_total_limit=early_stopping_patience + 1,
optim="adamw_torch",
)
# Remove experiment params from `training_args_kwargs`
for p in [
"output_dir",
"num_train_epochs",
"per_device_train_batch_size",
"per_device_eval_batch_size",
]:
if p in training_args_kwargs:
warnings.warn(
f"Argument {p} was passed in training_args_kwargs but will be ignored..."
)
training_args_kwargs.pop(p)
self.training_args_kwargs.update(training_args_kwargs)
self.train_size = train_size
self.mask_rate = mask_rate
self.columns: List[str] = []
self.column_dtypes: Dict[str, type] = {}
self.column_has_missing: Dict[str, bool] = {}
self.drop_na_cols: List[str] = []
self.processed_columns: List[str] = []
self.numeric_columns: List[str] = []
self.datetime_columns: List[str] = []
self.vocab: Dict[str, dict] = {}
# Output length for generator model
# including special tokens.
self.tabular_max_length = None
self.relational_max_length = None
# Number of derived columns for the relational
# and tabular data after performing the data transformation.
# This will be used as record size validator in the
# sampling stage.
self.tabular_col_size = None
self.relational_col_size = None
# This stores the transformation
# parameters for numeric columns.
self.col_transform_data: Optional[Dict] = None
# This is the col_transform_data
# for the relational models's in_df.
self.in_col_transform_data: Optional[Dict] = None
self.col_idx_ids: Dict[int, list] = {}
self.random_state = random_state
self.numeric_nparts = numeric_nparts
self.numeric_precision = numeric_precision
self.numeric_max_len = numeric_max_len
# A unique identifier for the experiment set after the
# model is trained.
self.experiment_id = None
self.trainer_state = None
# Target column, when set, a copy of the column values will be
# implicitly placed at the beginning of the dataframe.
self.target_col = None
self.realtabformer_version = realtabformer.__version__
[docs] def _invalid_model_type(self, model_type):
raise ValueError(
f"Model type: {model_type} is not valid. REaLTabFormer only supports \
`tabular` and `relational` values."
)
[docs] def _init_tabular(self, tabular_config):
if tabular_config is not None:
warnings.warn(
"The `bos_token_id`, `eos_token_id`, and `vocab_size` attributes will \
be replaced when the `.fit` method is run."
)
else:
# Default is 12, use 6 for distill-gpt2 as default
tabular_config = GPT2Config(n_layer=6)
self.tabular_config = tabular_config
self.model = None
[docs] def _init_relational(self, relational_config):
if relational_config is not None:
warnings.warn(
"The `bos_token_id`, `eos_token_id`, and `vocab_size` attributes for the \
encoder and decoder will be replaced when the `.fit` method is run."
)
else:
# Default is 12, use 6 for distill-gpt2 as default
relational_config = EncoderDecoderConfig(
encoder=GPT2Config(n_layer=6).to_dict(),
decoder=GPT2Config(n_layer=6).to_dict(),
)
if self.parent_gpt2_config is not None:
warnings.warn(
"A trained model for the parent table is available. The encoder will use the \
pretrained config and weights."
)
relational_config.encoder = GPT2Config(**self.parent_gpt2_config)
self.relational_config = relational_config
self.model = None
[docs] def _extract_column_info(self, df: pd.DataFrame) -> None:
# Track the column order of the original data
self.columns = df.columns.to_list()
# Store the dtypes of the columns
self.column_dtypes = df.dtypes.astype(str).to_dict()
# Track which variables have missing values
self.column_has_missing = (df.isnull().sum() > 0).to_dict()
# Get the columns where there should be no missing values
self.drop_na_cols = [
col for col, has_na in self.column_has_missing.items() if not has_na
]
# Identify the numeric columns. These will undergo
# special preprocessing.
self.numeric_columns = df.select_dtypes(include=np.number).columns.to_list()
# Identify the datetime columns. These will undergo
# special preprocessing.
self.datetime_columns = df.select_dtypes(include="datetime").columns.to_list()
[docs] def _generate_vocab(self, df: pd.DataFrame) -> dict:
return build_vocab(df, special_tokens=SpecialTokens.tokens(), add_columns=False)
[docs] def _check_model(self):
assert self.model is not None, "Model is None. Train the model first!"
[docs] def _split_train_eval_dataset(self, dataset: Dataset):
test_size = 1 - self.train_size
if test_size > 0:
dataset = dataset.train_test_split(
test_size=test_size, seed=self.random_state
)
dataset["train_dataset"] = dataset.pop("train")
dataset["eval_dataset"] = dataset.pop("test")
# Override `metric_for_best_model` from "loss" to "eval_loss"
self.training_args_kwargs["metric_for_best_model"] = "eval_loss"
# Make this explicit so that no assumption is made on the
# direction of the metric improvement.
self.training_args_kwargs["greater_is_better"] = False
else:
dataset = dict(train_dataset=dataset)
self.training_args_kwargs["eval_strategy"] = "no"
self.training_args_kwargs["load_best_model_at_end"] = False
return dataset
[docs] def fit(
self,
df: pd.DataFrame,
in_df: Optional[pd.DataFrame] = None,
join_on: Optional[str] = None,
resume_from_checkpoint: Union[bool, str] = False,
device="cuda",
num_bootstrap: int = 500,
frac: float = 0.165,
frac_max_data: int = 10000,
qt_max: Union[str, float] = 0.05,
qt_max_default: float = 0.05,
qt_interval: int = 100,
qt_interval_unique: int = 100,
distance: manhattan_distances = manhattan_distances,
quantile: float = 0.95,
n_critic: int = 5,
n_critic_stop: int = 2,
gen_rounds: int = 3,
sensitivity_max_col_nums: int = 20,
use_ks: bool = False,
full_sensitivity: bool = False,
sensitivity_orig_frac_multiple: int = 4,
orig_samples_rounds: int = 5,
load_from_best_mean_sensitivity: bool = False,
target_col: str = None,
):
"""Train the REaLTabFormer model on the tabular data.
Args:
df: Pandas DataFrame containing the tabular data that will be generated during sampling.
This data goes to the decoder for the relational model.
in_df: Pandas DataFrame containing observations related to `df`, and from which the
model will generate data. This data goes to the encoder for the relational model.
join_on: Column name that links the `df` and the `in_df` tables.
resume_from_checkpoint: If True, resumes training from the latest checkpoint in the
checkpoints_dir. If path, resumes the training from the given checkpoint.
device: Device where the model and the training will be run.
Use torch devices, e.g., `cpu`, `cuda`, `mps` (experimental)
num_bootstrap: Number of Bootstrap samples
frac: The fraction of the data used for training.
frac_max_data: The maximum number of rows that the training data will have.
qt_max: The maximum quantile for the discriminator.
qt_max_default: The default maximum quantile for the discriminator.
qt_interval: Interval for the quantile check during the training process.
qt_interval_unique: Interval for the quantile check during the training process.
distance: Distance metric used for discriminator.
quantile: The quantile value that the discriminator will be trained to.
n_critic: Interval between epochs to perform a discriminator assessment.
n_critic_stop: The number of critic rounds without improvement after which the training
will be stopped.
gen_rounds: The number of generator rounds.
sensitivity_max_col_nums: The maximum number of columns used to compute sensitivity.
use_ks: Whether to use KS test or not.
full_sensitivity: Whether to use full sensitivity or not.
sensitivity_orig_frac_multiple: The size of the training data relative to the chosen
`frac` that will be used in computing the sensitivity. The larger this value is, the
more robust the sensitivity threshold will be. However,
`(sensitivity_orig_frac_multiple + 2)` multiplied by `frac` must be less than 1.
orig_samples_rounds: This is the number of train/hold-out samples that will be used to
compute the epoch sensitivity value.
load_from_best_mean_sensitivity: Whether to load from best mean sensitivity or not.
target_col: The target column name.
Returns:
Trainer
"""
device = _validate_get_device(device)
# Set target col for teacher forcing
self.target_col = target_col
# Set the seed for, *hopefully*, replicability.
# This may cause an unexpected behavior when using
# the resume_from_checkpoint option.
if self.random_state:
random.seed(self.random_state)
np.random.seed(self.random_state)
torch.manual_seed(self.random_state)
torch.cuda.manual_seed_all(self.random_state)
if self.model_type == ModelType.tabular:
if n_critic <= 0:
trainer = self._fit_tabular(df, device=device)
trainer.train(resume_from_checkpoint=resume_from_checkpoint)
else:
trainer = self._train_with_sensitivity(
df,
device,
num_bootstrap=num_bootstrap,
frac=frac,
frac_max_data=frac_max_data,
qt_max=qt_max,
qt_max_default=qt_max_default,
qt_interval=qt_interval,
qt_interval_unique=qt_interval_unique,
distance=distance,
quantile=quantile,
n_critic=n_critic,
n_critic_stop=n_critic_stop,
gen_rounds=gen_rounds,
resume_from_checkpoint=resume_from_checkpoint,
sensitivity_max_col_nums=sensitivity_max_col_nums,
use_ks=use_ks,
full_sensitivity=full_sensitivity,
sensitivity_orig_frac_multiple=sensitivity_orig_frac_multiple,
orig_samples_rounds=orig_samples_rounds,
load_from_best_mean_sensitivity=load_from_best_mean_sensitivity,
)
del self.dataset
elif self.model_type == ModelType.relational:
assert (
in_df is not None
), "The REaLTabFormer for relational data requires two tables for training."
assert join_on is not None, "The column to join the data must not be None."
trainer = self._fit_relational(df, in_df, join_on=join_on, device=device)
trainer.train(resume_from_checkpoint=resume_from_checkpoint)
else:
self._invalid_model_type(self.model_type)
try:
self.experiment_id = f"id{int((time.time() * 10 ** 10)):024}"
torch.cuda.empty_cache()
return trainer
except Exception as exception:
if device == torch.device("cuda"):
del self.model
torch.cuda.empty_cache()
self.model = None
raise exception
[docs] def _train_with_sensitivity(
self,
df: pd.DataFrame,
device: str = "cuda",
num_bootstrap: int = 500,
frac: float = 0.165,
frac_max_data: int = 10000,
qt_max: Union[str, float] = 0.05,
qt_max_default: float = 0.05,
qt_interval: int = 100,
qt_interval_unique: int = 100,
distance: manhattan_distances = manhattan_distances,
quantile: float = 0.95,
n_critic: int = 5,
n_critic_stop: int = 2,
gen_rounds: int = 3,
sensitivity_max_col_nums: int = 20,
use_ks: bool = False,
resume_from_checkpoint: Union[bool, str] = False,
full_sensitivity: bool = False,
sensitivity_orig_frac_multiple: int = 4,
orig_samples_rounds: int = 5,
load_from_best_mean_sensitivity: bool = False,
):
assert gen_rounds >= 1
_frac = min(frac, frac_max_data / len(df))
if frac != _frac:
warnings.warn(
f"The frac ({frac}) set results to a sample larger than \
frac_max_data={frac_max_data}. Setting frac to {_frac}."
)
frac = _frac
trainer: Trainer = None
dup_rate = df.duplicated().mean()
if isinstance(qt_max, str):
if qt_max == "compute":
# The idea behind this is if the empirical has
# natural duplicates, we can use that as
# basis for what a typical rate for duplicates a
# random sample should have. Any signidican excess
# from this indicates overfitting.
# The choice of dividing the duplicate rate by 2
# is arbitrary but reasonable to prevent delayed
# stopping when overfitting.
dup_rate = dup_rate / 2
qt_max = dup_rate if dup_rate > 0 else qt_max_default
else:
raise ValueError(f"Unexpected qt_max value: {qt_max}")
elif not isinstance(qt_max, str) and dup_rate >= qt_max:
warnings.warn(
f'The qt_max ({qt_max}) set is lower than the duplicate \rate ({dup_rate}) in \
the data. This will not give a reliable early stopping condition. Consider \
using qt_max="compute" argument.'
)
if dup_rate == 0:
# We do this because for data without unique values, we
# expect that a generated sample should have equal likelihood
# in the minimum distance with the hold out.
warnings.warn(
f"Duplicate rate ({dup_rate}) in the data is zero. The `qt_interval` will be set \
to qt_interval_unique={qt_interval_unique}."
)
qt_interval = qt_interval_unique
# Estimate the sensitivity threshold
print("Computing the sensitivity threshold...")
if not full_sensitivity:
# Dynamically compute the qt_interval to fit the data
# if the resulting sample has lower resolution.
# For example, we can't use qt_interval=1000 if the number
# of samples left at qt_max of the distance matrix is less than
# 1000.
# The formula means:
# - 2 -> accounts for the fact that we concatenate the rows and columns
# of the distance matrix.
# - frac -> the proportion of the training data that is used to compute the
# the distance matrix.
# - qt_max -> the maximum quantile of assessment.
# We divide by 2 to increase the resolution a bit
_qt_interval = min(qt_interval, (2 * frac * len(df) * qt_max) // 2)
_qt_interval = max(_qt_interval, 2)
_qt_interval = int(_qt_interval)
if _qt_interval < qt_interval:
warnings.warn(
f"qt_interval adjusted from {qt_interval} to {_qt_interval}..."
)
qt_interval = _qt_interval
# Computing this here before splitting may have some data
# leakage issue, but it should be almost negligible. Doing
# the computation of the threshold on the full data with the
# train size aligned will give a more reliable estimate of
# the sensitivity threshold.
sensitivity_values = SyntheticDataBench.compute_sensitivity_threshold(
train_data=df,
num_bootstrap=num_bootstrap,
# Divide by two so that the train_data in this computation matches the size
# of the final df used to train the model. This is essential so that the
# sensitivity_threshold value is consistent with the val_sensitivity.
# Concretely, the computation of the distribution of min distances is
# relative to the number of training observations.
# The `frac` in this method corresponds to the size of both the test and the
# synthetic samples.
frac=frac / 2,
qt_max=qt_max,
qt_interval=qt_interval,
distance=distance,
return_values=True,
quantile=quantile,
max_col_nums=sensitivity_max_col_nums,
use_ks=use_ks,
full_sensitivity=full_sensitivity,
sensitivity_orig_frac_multiple=sensitivity_orig_frac_multiple,
)
sensitivity_threshold = np.quantile(sensitivity_values, quantile)
mean_sensitivity_value = np.mean(sensitivity_values)
best_mean_sensitivity_value = np.inf
assert isinstance(sensitivity_threshold, float)
print("Sensitivity threshold:", sensitivity_threshold, "qt_max:", qt_max)
# # Create a hold out sample for the discriminator model
# hold_df = df.sample(frac=frac, random_state=self.random_state)
# df = df.loc[df.index.difference(hold_df.index)]
# Start training
logging.info("Start training...")
# Remove existing checkpoints
for chkp in self.checkpoints_dir.glob("checkpoint-*"):
shutil.rmtree(chkp, ignore_errors=True)
sensitivity_scores = []
bdm_path = self.checkpoints_dir / TabularArtefact.best_disc_model
mean_closest_bdm_path = (
self.checkpoints_dir / TabularArtefact.mean_best_disc_model
)
not_bdm_path = self.checkpoints_dir / TabularArtefact.not_best_disc_model
last_epoch_path = self.checkpoints_dir / TabularArtefact.last_epoch_model
# Remove existing artefacts in the best model dir
shutil.rmtree(bdm_path, ignore_errors=True)
bdm_path.mkdir(parents=True, exist_ok=True)
shutil.rmtree(mean_closest_bdm_path, ignore_errors=True)
mean_closest_bdm_path.mkdir(parents=True, exist_ok=True)
shutil.rmtree(not_bdm_path, ignore_errors=True)
not_bdm_path.mkdir(parents=True, exist_ok=True)
shutil.rmtree(last_epoch_path, ignore_errors=True)
last_epoch_path.mkdir(parents=True, exist_ok=True)
last_epoch = 0
not_best_val_sensitivity = np.inf
if resume_from_checkpoint:
chkp_list = sorted(
self.checkpoints_dir.glob("checkpoint-*"), key=os.path.getmtime
)
if chkp_list:
# Get the most recent checkpoint based on
# creation time.
chkp = chkp_list[-1]
trainer_state = json.loads((chkp / "trainer_state.json").read_text())
last_epoch = math.ceil(trainer_state["epoch"])
trainer = self._fit_tabular(
df,
device=device,
num_train_epochs=last_epoch,
target_epochs=self.epochs,
)
np.random.seed(self.random_state)
random.seed(self.random_state)
for p_epoch in range(last_epoch, self.epochs, n_critic):
gen_total = int(len(df) * frac)
num_train_epochs = min(p_epoch + n_critic, self.epochs)
# Perform the discriminator sampling every `n_critic` epochs
if trainer is None:
trainer = self._fit_tabular(
df,
device=device,
num_train_epochs=num_train_epochs,
target_epochs=self.epochs,
)
trainer.train(resume_from_checkpoint=False)
else:
trainer = self._build_tabular_trainer(
device=device,
num_train_epochs=num_train_epochs,
target_epochs=self.epochs,
)
trainer.train(resume_from_checkpoint=True)
try:
# Generate samples
gen_df = self.sample(n_samples=gen_rounds * gen_total, device=device)
except SampleEmptyLimitError:
# Continue training if the model is still not
# able to generate stable observations.
continue
val_sensitivities = []
if full_sensitivity:
for _ in range(gen_rounds):
hold_df = df.sample(n=gen_total)
for g_idx in range(gen_rounds):
val_sensitivities.append(
SyntheticDataBench.compute_sensitivity_metric(
original=df.loc[df.index.difference(hold_df.index)],
synthetic=gen_df.iloc[
g_idx * gen_total : (g_idx + 1) * gen_total
],
test=hold_df,
qt_max=qt_max,
qt_interval=qt_interval,
distance=distance,
max_col_nums=sensitivity_max_col_nums,
use_ks=use_ks,
)
)
else:
for g_idx in range(gen_rounds):
for _ in range(orig_samples_rounds):
original_df = df.sample(
n=sensitivity_orig_frac_multiple * gen_total, replace=False
)
hold_df = df.loc[df.index.difference(original_df.index)].sample(
n=gen_total, replace=False
)
val_sensitivities.append(
SyntheticDataBench.compute_sensitivity_metric(
original=original_df,
synthetic=gen_df.iloc[
g_idx * gen_total : (g_idx + 1) * gen_total
],
test=hold_df,
qt_max=qt_max,
qt_interval=qt_interval,
distance=distance,
max_col_nums=sensitivity_max_col_nums,
use_ks=use_ks,
)
)
val_sensitivity = np.mean(val_sensitivities)
sensitivity_scores.append(val_sensitivity)
if val_sensitivity < sensitivity_threshold:
# Just save the model while the
# validation sensitivity is still within
# the accepted range.
# This way we can load the acceptable
# model back when the threshold is breached.
trainer.save_model(bdm_path.as_posix())
trainer.state.save_to_json((bdm_path / "trainer_state.json").as_posix())
elif not_best_val_sensitivity > (val_sensitivity - sensitivity_threshold):
print("Saving not-best model...")
trainer.save_model(not_bdm_path.as_posix())
trainer.state.save_to_json(
(not_bdm_path / "trainer_state.json").as_posix()
)
not_best_val_sensitivity = val_sensitivity - sensitivity_threshold
_delta_mean_sensitivity_value = abs(
mean_sensitivity_value - val_sensitivity
)
if _delta_mean_sensitivity_value < best_mean_sensitivity_value:
best_mean_sensitivity_value = _delta_mean_sensitivity_value
trainer.save_model(mean_closest_bdm_path.as_posix())
trainer.state.save_to_json(
(mean_closest_bdm_path / "trainer_state.json").as_posix()
)
print(
f"Critic round: {p_epoch + n_critic}, \
sensitivity_threshold: {sensitivity_threshold}, \
val_sensitivity: {val_sensitivity}, \
val_sensitivities: {val_sensitivities}"
)
if len(sensitivity_scores) > n_critic_stop:
n_no_improve = 0
for sensitivity_score in sensitivity_scores[-n_critic_stop:]:
# We count no improvement if the score is not
# better than the best, and that the score is not
# better than the previous score.
if sensitivity_score > sensitivity_threshold:
n_no_improve += 1
if n_no_improve == n_critic_stop:
print("Stopping training, no improvement in critic...")
break
# Save last epoch artefacts before loading the best model.
trainer.save_model(last_epoch_path.as_posix())
trainer.state.save_to_json((last_epoch_path / "trainer_state.json").as_posix())
loaded_model_path = None
if not load_from_best_mean_sensitivity:
if (bdm_path / "pytorch_model.bin").exists() or (bdm_path / "model.safetensors").exists():
loaded_model_path = bdm_path
else:
if (mean_closest_bdm_path / "pytorch_model.bin").exists() or (mean_closest_bdm_path / "model.safetensors").exists():
loaded_model_path = mean_closest_bdm_path
if loaded_model_path is None:
# There should always be at least one `mean_closest_bdm_path` but
# in case it doesn't exist, try loading from `not_bdm_path`.
warnings.warn(
"No best model was saved. Loading the closest model to the sensitivity_threshold."
)
loaded_model_path = not_bdm_path
self.model = self.model.from_pretrained(loaded_model_path.as_posix())
self.trainer_state = json.loads(
(loaded_model_path / "trainer_state.json").read_text()
)
return trainer
[docs] def _set_up_relational_coder_configs(self) -> None:
def _get_coder(coder_name) -> GPT2Config:
return getattr(self.relational_config, coder_name)
for coder_name in ["encoder", "decoder"]:
coder = _get_coder(coder_name)
coder.bos_token_id = self.vocab[coder_name]["token2id"][SpecialTokens.BOS]
coder.eos_token_id = self.vocab[coder_name]["token2id"][SpecialTokens.EOS]
coder.pad_token_id = self.vocab[coder_name]["token2id"][SpecialTokens.PAD]
coder.vocab_size = len(self.vocab[coder_name]["id2token"])
if coder_name == "decoder":
self.relational_config.bos_token_id = coder.bos_token_id
self.relational_config.eos_token_id = coder.eos_token_id
self.relational_config.pad_token_id = coder.pad_token_id
self.relational_config.decoder_start_token_id = coder.eos_token_id
# Make sure that we have at least the number of
# columns in the transformed data as positions.
# This will prevent runtime error.
# `RuntimeError: CUDA error: device-side assert triggered`
assert self.relational_max_length
if (
coder_name == "decoder"
and coder.n_positions < self.relational_max_length
):
coder.n_positions = 128 + self.relational_max_length
elif coder_name == "encoder" and getattr(
coder, "n_positions", getattr(coder, "max_position_embeddings")
) < len(self.vocab[coder_name]["column_token_ids"]):
positions = 128 + len(self.vocab[coder_name]["column_token_ids"])
try:
coder.n_positions = positions
except:
coder.max_position_embeddings = positions
# This must be set to True for the EncoderDecoderModel to work at least
# with GPT2 as the decoder.
self.relational_config.decoder.add_cross_attention = True
[docs] def _fit_relational(
self, out_df: pd.DataFrame, in_df: pd.DataFrame, join_on: str, device="cuda"
):
# bert2bert.config.decoder_start_token_id = tokenizer.cls_token_id
# bert2bert.config.eos_token_id = tokenizer.sep_token_id
# bert2bert.config.pad_token_id = tokenizer.pad_token_id
# bert2bert.config.vocab_size = bert2bert.config.encoder.vocab_size
# All join values in the out_df must be present in the in_df.
assert len(set(out_df[join_on].unique()).difference(in_df[join_on])) == 0
# Get the list of index of observations that are related based on
# the join_on variable.
common_out_idx = (
out_df.reset_index(drop=True)
.groupby(join_on)
.apply(lambda x: x.index.to_list())
)
# Track the mapping of index from input to the list of output indices.
in_out_idx = pd.Series(
# Reset the index so that we are sure that the index ids are set properly.
dict(in_df[join_on].reset_index(drop=True).items())
).map(lambda x: common_out_idx.get(x, []))
# Remove the unique id column from the in_df and the out_df
in_df = in_df.drop(join_on, axis=1)
out_df = out_df.drop(join_on, axis=1)
self._extract_column_info(out_df)
out_df, self.col_transform_data = process_data(
out_df,
numeric_max_len=self.numeric_max_len,
numeric_precision=self.numeric_precision,
numeric_nparts=self.numeric_nparts,
)
self.processed_columns = out_df.columns.to_list()
self.vocab["decoder"] = self._generate_vocab(out_df)
self.relational_col_size = out_df.shape[1]
# NOTE: the index starts at zero, but should be adjusted
# to account for the special tokens. For relational data,
# the index should start at 3 ([[EOS], [BOS], [BMEM]]).
self.col_idx_ids = {
ix: self.vocab["decoder"]["column_token_ids"][col]
for ix, col in enumerate(self.processed_columns)
}
# Add these special tokens at specific key values
# which are used in `REaLSampler._get_relational_col_idx_ids`
self.col_idx_ids[-1] = [
self.vocab["decoder"]["token2id"][SpecialTokens.BMEM],
self.vocab["decoder"]["token2id"][SpecialTokens.EOS],
]
self.col_idx_ids[-2] = [self.vocab["decoder"]["token2id"][SpecialTokens.EMEM]]
# TODO: handle the col_transform_data from the in_df as well.
in_df, self.in_col_transform_data = process_data(
in_df,
numeric_max_len=self.numeric_max_len,
numeric_precision=self.numeric_precision,
numeric_nparts=self.numeric_nparts,
col_transform_data=self.parent_col_transform_data,
)
if self.parent_vocab is None:
self.vocab["encoder"] = self._generate_vocab(in_df)
else:
self.vocab["encoder"] = self.parent_vocab
# Load the dataframe into a HuggingFace Dataset
dataset = make_relational_dataset(
in_df=in_df,
out_df=out_df,
vocab=self.vocab,
in_out_idx=in_out_idx,
output_max_length=self.output_max_length,
mask_rate=self.mask_rate,
return_token_type_ids=False,
)
# Compute the longest sequence of labels in the dataset and add a buffer of 1.
self.relational_max_length = (
max(
dataset.map(lambda example: dict(length=len(example["labels"])))[
"length"
]
)
+ 1
)
# Create train-eval split if specified
dataset = self._split_train_eval_dataset(dataset)
# Set up the config and the model
self._set_up_relational_coder_configs()
# Build the model.
self.model = EncoderDecoderModel(self.relational_config)
if self.parent_gpt2_state_dict is not None:
pretrain_load = self.model.encoder.load_state_dict(
self.parent_gpt2_state_dict, strict=False
)
assert (
not pretrain_load.missing_keys
), "There should be no missing_keys after loading the pretrained GPT2 state!"
if self.freeze_parent_model:
# We freeze the weights if we use the pretrained
# parent table model.
for param in self.model.encoder.parameters():
param.requires_grad = False
# Tell pytorch to run this model on the GPU.
device = torch.device(device)
if device == torch.device("cuda"):
self.model.cuda()
# Set TrainingArguments and the Seq2SeqTrainer
training_args_kwargs = dict(self.training_args_kwargs)
default_args_kwargs = dict(
# predict_with_generate=True,
# warmup_steps=2000,
fp16=(
device == torch.device("cuda")
), # Use fp16 by default if using cuda device
)
for k, v in default_args_kwargs.items():
if k not in training_args_kwargs:
training_args_kwargs[k] = v
callbacks = None
if training_args_kwargs["load_best_model_at_end"]:
callbacks = [
EarlyStoppingCallback(
self.early_stopping_patience, self.early_stopping_threshold
)
]
# instantiate trainer
trainer = Seq2SeqTrainer(
model=self.model,
args=Seq2SeqTrainingArguments(**training_args_kwargs),
callbacks=callbacks,
data_collator=RelationalDataCollator(),
**dataset,
)
return trainer
[docs] def _fit_tabular(
self,
df: pd.DataFrame,
device="cuda",
num_train_epochs: int = None,
target_epochs: int = None,
) -> Trainer:
self._extract_column_info(df)
df, self.col_transform_data = process_data(
df,
numeric_max_len=self.numeric_max_len,
numeric_precision=self.numeric_precision,
numeric_nparts=self.numeric_nparts,
target_col=self.target_col,
)
self.processed_columns = df.columns.to_list()
self.vocab = self._generate_vocab(df)
self.tabular_col_size = df.shape[0]
# NOTE: the index starts at zero, but should be adjusted
# to account for the special tokens. For tabular data,
# the index should start at 1.
self.col_idx_ids = {
ix: self.vocab["column_token_ids"][col]
for ix, col in enumerate(self.processed_columns)
}
# Load the dataframe into a HuggingFace Dataset
dataset = make_dataset(
df, self.vocab, mask_rate=self.mask_rate, return_token_type_ids=False
)
# Store the sequence length for the processed data
self.tabular_max_length = len(dataset[0]["input_ids"])
# Create train-eval split if specified
dataset = self._split_train_eval_dataset(dataset)
self.dataset = dataset
# Set up the config and the model
self.tabular_config.bos_token_id = self.vocab["token2id"][SpecialTokens.BOS]
self.tabular_config.eos_token_id = self.vocab["token2id"][SpecialTokens.EOS]
self.tabular_config.vocab_size = len(self.vocab["id2token"])
# Make sure that we have at least the number of
# columns in the transformed data as positions.
if self.tabular_config.n_positions < len(self.vocab["column_token_ids"]):
self.tabular_config.n_positions = 128 + len(self.vocab["column_token_ids"])
self.model = GPT2LMHeadModel(self.tabular_config)
# Tell pytorch to run this model on the GPU.
device = torch.device(device)
if device == torch.device("cuda"):
self.model.cuda()
return self._build_tabular_trainer(
device=device,
num_train_epochs=num_train_epochs,
target_epochs=target_epochs,
)
[docs] def _build_tabular_trainer(
self,
device="cuda",
num_train_epochs: int = None,
target_epochs: int = None,
) -> Trainer:
device = torch.device(device)
# Set TrainingArguments and the Trainer
logging.info("Set up the TrainingArguments and the Trainer...")
training_args_kwargs: Dict[str, Any] = dict(self.training_args_kwargs)
default_args_kwargs = dict(
fp16=(
device == torch.device("cuda")
), # Use fp16 by default if using cuda device
)
for k, v in default_args_kwargs.items():
if k not in training_args_kwargs:
training_args_kwargs[k] = v
if num_train_epochs is not None:
training_args_kwargs["num_train_epochs"] = num_train_epochs
# # NOTE: The `ResumableTrainer` will default to its original
# # behavior (Trainer) if `target_epochs`` is None.
# # Set the `target_epochs` to `num_train_epochs` if not specified.
# if target_epochs is None:
# target_epochs = training_args_kwargs.get("num_train_epochs")
callbacks = None
if training_args_kwargs["load_best_model_at_end"]:
callbacks = [
EarlyStoppingCallback(
self.early_stopping_patience, self.early_stopping_threshold
)
]
assert self.dataset
trainer = ResumableTrainer(
target_epochs=target_epochs,
save_epochs=None,
model=self.model,
args=TrainingArguments(**training_args_kwargs),
data_collator=None, # Use the default_data_collator
callbacks=callbacks,
**self.dataset,
)
return trainer
[docs] def sample(
self,
n_samples: int = None,
input_unique_ids: Optional[Union[pd.Series, List]] = None,
input_df: Optional[pd.DataFrame] = None,
input_ids: Optional[torch.tensor] = None,
gen_batch: Optional[int] = 128,
device: str = "cuda",
seed_input: Optional[Union[pd.DataFrame, Dict[str, Any]]] = None,
save_samples: Optional[bool] = False,
constrain_tokens_gen: Optional[bool] = True,
validator: Optional[ObservationValidator] = None,
continuous_empty_limit: int = 10,
suppress_tokens: Optional[List[int]] = None,
forced_decoder_ids: Optional[List[List[int]]] = None,
related_num: Optional[Union[int, List[int]]] = None,
**generate_kwargs,
) -> pd.DataFrame:
"""Generate synthetic tabular data samples
Args:
n_samples: Number of synthetic samples to generate for the tabular data.
input_unique_ids: The unique identifier that will be used to link the input
data to the generated values when sampling for relational data.
input_df: Pandas DataFrame containing the tabular input data.
input_ids: (NOTE: the `input_df` argument is the preferred input)
The input_ids that conditions the generation of the relational data.
gen_batch: Controls the batch size of the data generation process. This parameter
should be adjusted based on the compute resources.
device: The device used by the generator.
Use torch devices, e.g., `cpu`, `cuda`, `mps` (experimental)
seed_input: A dictionary of `col_name:values` for the seed data. Only `col_names`
that are actually in the first sequence of the training input will be used.
constrain_tokens_gen: Set whether we impose a constraint at each step of the generation
limited only to valid tokens for the column.
validator: An instance of `ObservationValidator` for validating the generated samples.
The validators are applied to observations only, and don't support inter-observation
validation. See `ObservationValidator` docs on how to set up a validator.
continuous_invalid_limit: The sampling will raise an exception if
`continuous_empty_limit` empty sample batches have been produced continuously. This
will prevent an infinite loop if the quality of the data generated is not good and
always produces invalid observations.
suppress_tokens: (from docs) A list of tokens that will be supressed at generation.
The SupressTokens logit processor will set their log probs to -inf so that they are
not sampled. This is a useful feature for imputing missing values.
forced_decoder_ids: (from docs) A list of pairs of integers which indicates a mapping
from generation indices to token indices that will be forced before sampling. For
example, [[1, 123]] means the second generated token will always be a token of
index 123. This is a useful feature for constraining the model to generate only
specific stratification variables in surveys, e.g., GEO1, URBAN/RURAL variables.
related_num: A column name in the input_df containing the number of observations that the child
table is expected to have for the parent observation. It can also be an integer if the input_df
corresponds to a set of observations having the same number of expected observations.
This parameter is only valid for the relational model.
generate_kwargs: Additional keywords arguments that will be supplied to `.generate`
method. For a comprehensive list of arguments, see:
https://huggingface.co/docs/transformers/v4.24.0/en/main_classes/text_generation#transformers.generation_utils.GenerationMixin.generate
Returns:
DataFrame with n_samples rows of generated data
"""
self._check_model()
device = _validate_get_device(device)
# Clear the cache
torch.cuda.empty_cache()
if self.model_type == ModelType.tabular:
assert n_samples
assert self.tabular_max_length is not None
assert self.tabular_col_size is not None
assert self.col_transform_data is not None
tabular_sampler = TabularSampler.sampler_from_model(
rtf_model=self, device=device
)
# (
# model_type=self.model_type,
# model=self.model,
# vocab=self.vocab,
# processed_columns=self.processed_columns,
# max_length=self.tabular_max_length,
# col_size=self.tabular_col_size,
# col_idx_ids=self.col_idx_ids,
# columns=self.columns,
# datetime_columns=self.datetime_columns,
# column_dtypes=self.column_dtypes,
# drop_na_cols=self.drop_na_cols,
# col_transform_data=self.col_transform_data,
# random_state=self.random_state,
# device=device,
# )
synth_df = tabular_sampler.sample_tabular(
n_samples=n_samples,
gen_batch=gen_batch,
device=device,
seed_input=seed_input,
constrain_tokens_gen=constrain_tokens_gen,
validator=validator,
continuous_empty_limit=continuous_empty_limit,
suppress_tokens=suppress_tokens,
forced_decoder_ids=forced_decoder_ids,
**generate_kwargs,
)
elif self.model_type == ModelType.relational:
assert (input_ids is not None) or (input_df is not None)
assert self.relational_max_length is not None
assert self.relational_col_size is not None
assert self.col_transform_data is not None
assert self.in_col_transform_data is not None
relational_sampler = RelationalSampler.sampler_from_model(
rtf_model=self, device=device
)
# (
# model_type=self.model_type,
# model=self.model,
# vocab=self.vocab,
# processed_columns=self.processed_columns,
# max_length=self.relational_max_length,
# col_size=self.relational_col_size,
# col_idx_ids=self.col_idx_ids,
# columns=self.columns,
# datetime_columns=self.datetime_columns,
# column_dtypes=self.column_dtypes,
# drop_na_cols=self.drop_na_cols,
# col_transform_data=self.col_transform_data,
# in_col_transform_data=self.in_col_transform_data,
# random_state=self.random_state,
# device=device,
# )
synth_df = relational_sampler.sample_relational(
input_unique_ids=input_unique_ids,
input_df=input_df,
input_ids=input_ids,
device=device,
gen_batch=gen_batch,
constrain_tokens_gen=constrain_tokens_gen,
validator=validator,
continuous_empty_limit=continuous_empty_limit,
suppress_tokens=suppress_tokens,
forced_decoder_ids=forced_decoder_ids,
related_num=related_num,
**generate_kwargs,
)
if save_samples:
samples_fname = (
self.samples_save_dir
/ f"rtf_{self.model_type}-exp_{self.experiment_id}-{int(time.time())}-samples_{synth_df.shape[0]}.pkl"
)
samples_fname.parent.mkdir(parents=True, exist_ok=True)
synth_df.to_pickle(samples_fname)
return synth_df
[docs] def predict(
self,
data: pd.DataFrame,
target_col: str,
target_pos_val: Any = None,
batch: int = 32,
obs_sample: int = 30,
fillunk: bool = True,
device: str = "cuda",
disable_progress_bar: bool = True,
**generate_kwargs,
) -> pd.Series:
"""
Use the trained model to make predictions on a given dataframe.
Args:
data: The data to make predictions on, in the form of a Pandas dataframe.
target_col: The name of the target column in the data to predict.
target_pos_val: The positive value in the target column to use for binary
classification. This is produces a one-to-many prediction relative to
`target_pos_val` for targets that are multi-categorical.
batch: The batch size to use when making predictions.
obs_sample: The number of observations to sample from the data when making predictions.
fillunk: If True, the function will fill any missing values in the data before making
predictions. Fill unknown tokens with the mode of the batch in the given step.
device: The device to use for prediction. Can be either "cpu" or "cuda".
**generate_kwargs: Additional keyword arguments to pass to the model's `generate`
method.
Returns:
A Pandas series containing the predicted values for the target column.
"""
assert (
self.model_type == ModelType.tabular
), "The predict method is only implemented for tabular data..."
self._check_model()
device = _validate_get_device(device)
batch = min(batch, data.shape[0])
# Clear the cache
torch.cuda.empty_cache()
# assert self.tabular_max_length is not None
# assert self.tabular_col_size is not None
# assert self.col_transform_data is not None
tabular_sampler = TabularSampler.sampler_from_model(self, device=device)
# TabularSampler(
# model_type=self.model_type,
# model=self.model,
# vocab=self.vocab,
# processed_columns=self.processed_columns,
# max_length=self.tabular_max_length,
# col_size=self.tabular_col_size,
# col_idx_ids=self.col_idx_ids,
# columns=self.columns,
# datetime_columns=self.datetime_columns,
# column_dtypes=self.column_dtypes,
# drop_na_cols=self.drop_na_cols,
# col_transform_data=self.col_transform_data,
# random_state=self.random_state,
# device=device,
# )
return tabular_sampler.predict(
data=data,
target_col=target_col,
target_pos_val=target_pos_val,
batch=batch,
obs_sample=obs_sample,
fillunk=fillunk,
device=device,
disable_progress_bar=disable_progress_bar,
**generate_kwargs,
)
[docs] def save(self, path: Union[str, Path], allow_overwrite: Optional[bool] = False):
"""Save REaLTabFormer Model
Saves the model weights and a configuration file in the given directory.
Args:
path: Path where to save the model
"""
self._check_model()
assert self.experiment_id is not None
if isinstance(path, str):
path = Path(path)
# Add experiment id to the save path
path = path / self.experiment_id
config_file = path / ModelFileName.rtf_config_json
model_file = path / ModelFileName.rtf_model_pt
if path.is_dir() and not allow_overwrite:
if config_file.exists() or model_file.exists():
raise ValueError(
"This directory is not empty, and contains either a config or a model."
" Consider setting `allow_overwrite=True` if you want to overwrite these."
)
else:
warnings.warn(
f"Directory {path} exists, but `allow_overwrite=False`."
" This will raise an error next time when the model artifacts \
exist on this directory"
)
path.mkdir(parents=True, exist_ok=True)
# Save attributes
rtf_attrs = self.__dict__.copy()
rtf_attrs.pop("model")
# We don't need to store the `parent_config`
# since a saved model should have the weights loaded from
# the trained model already.
for ignore_key in [
"parent_vocab",
"parent_gpt2_config",
"parent_gpt2_state_dict",
"parent_col_transform_data",
]:
if ignore_key in rtf_attrs:
rtf_attrs.pop(ignore_key)
# GPT2Config is not JSON serializable, let us manually
# extract the attributes.
if rtf_attrs.get("tabular_config"):
rtf_attrs["tabular_config"] = rtf_attrs["tabular_config"].to_dict()
if rtf_attrs.get("relational_config"):
rtf_attrs["relational_config"] = rtf_attrs["relational_config"].to_dict()
rtf_attrs["checkpoints_dir"] = rtf_attrs["checkpoints_dir"].as_posix()
rtf_attrs["samples_save_dir"] = rtf_attrs["samples_save_dir"].as_posix()
config_file.write_text(json.dumps(rtf_attrs))
# Save model weights
torch.save(self.model.state_dict(), model_file.as_posix())
if self.model_type == ModelType.tabular:
# Copy the special model checkpoints for
# tabular models.
for artefact in TabularArtefact.artefacts():
print("Copying artefacts from:", artefact)
if (self.checkpoints_dir / artefact).exists():
shutil.copytree(
self.checkpoints_dir / artefact,
path / artefact,
dirs_exist_ok=True,
)
@classmethod
[docs] def load_from_dir(cls, path: Union[str, Path]):
"""Load a saved REaLTabFormer model
Load trained REaLTabFormer model from directory.
Args:
path: Directory where REaLTabFormer model is saved
Returns:
REaLTabFormer instance
"""
if isinstance(path, str):
path = Path(path)
config_file = path / ModelFileName.rtf_config_json
model_file = path / ModelFileName.rtf_model_pt
assert path.is_dir(), f"Directory {path} does not exist."
assert config_file.exists(), f"Config file {config_file} does not exist."
assert model_file.exists(), f"Model file {model_file} does not exist."
# Load the saved attributes
rtf_attrs = json.loads(config_file.read_text())
# Create new REaLTabFormer model instance
try:
realtf = cls(model_type=rtf_attrs["model_type"])
except KeyError:
# Back-compatibility for saved models
# before the support for relational data
# was implemented.
realtf = cls(model_type="tabular")
# Set all attributes and handle the
# special case for the GPT2Config.
for k, v in rtf_attrs.items():
if k == "gpt_config":
# Back-compatibility for saved models
# before the support for relational data
# was implemented.
v = GPT2Config.from_dict(v)
k = "tabular_config"
elif k == "tabular_config":
v = GPT2Config.from_dict(v)
elif k == "relational_config":
v = EncoderDecoderConfig.from_dict(v)
elif k in ["checkpoints_dir", "samples_save_dir"]:
v = Path(v)
elif k == "vocab":
if realtf.model_type == ModelType.tabular:
# Cast id back to int since JSON converts them to string.
v["id2token"] = {int(ii): vv for ii, vv in v["id2token"].items()}
elif realtf.model_type == ModelType.relational:
v["encoder"]["id2token"] = {
int(ii): vv for ii, vv in v["encoder"]["id2token"].items()
}
v["decoder"]["id2token"] = {
int(ii): vv for ii, vv in v["decoder"]["id2token"].items()
}
else:
raise ValueError(f"Invalid model_type: {realtf.model_type}")
elif k == "col_idx_ids":
v = {int(ii): vv for ii, vv in v.items()}
setattr(realtf, k, v)
# Implement back-compatibility for REaLTabFormer version < 0.0.1.8.2
# since the attribute `col_idx_ids` is not implemented before.
if "col_idx_ids" not in rtf_attrs:
if realtf.model_type == ModelType.tabular:
realtf.col_idx_ids = {
ix: realtf.vocab["column_token_ids"][col]
for ix, col in enumerate(realtf.processed_columns)
}
elif realtf.model_type == ModelType.relational:
# NOTE: the index starts at zero, but should be adjusted
# to account for the special tokens. For relational data,
# the index should start at 3 ([[EOS], [BOS], [BMEM]]).
realtf.col_idx_ids = {
ix: realtf.vocab["decoder"]["column_token_ids"][col]
for ix, col in enumerate(realtf.processed_columns)
}
# Add these special tokens at specific key values
# which are used in `REaLSampler._get_relational_col_idx_ids`
realtf.col_idx_ids[-1] = [
realtf.vocab["decoder"]["token2id"][SpecialTokens.BMEM],
realtf.vocab["decoder"]["token2id"][SpecialTokens.EOS],
]
realtf.col_idx_ids[-2] = [
realtf.vocab["decoder"]["token2id"][SpecialTokens.EMEM]
]
# Load model weights
if realtf.model_type == ModelType.tabular:
realtf.model = GPT2LMHeadModel(realtf.tabular_config)
elif realtf.model_type == ModelType.relational:
realtf.model = EncoderDecoderModel(realtf.relational_config)
else:
raise ValueError(f"Invalid model_type: {realtf.model_type}")
realtf.model.load_state_dict(
torch.load(model_file.as_posix(), map_location="cpu")
)
return realtf